Abstract
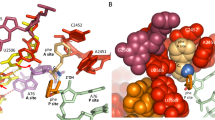
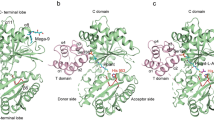
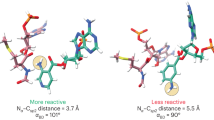
The large ribosomal subunit catalyses the reaction between the α-amino group of the aminoacyl-tRNA bound to the A site and the ester carbon of the peptidyl-tRNA bound to the P site1, while preventing the nucleophilic attack of water on the ester, which would lead to unprogrammed deacylation of the peptidyl-tRNA. Here we describe three new structures of the large ribosomal subunit of Haloarcula marismortui (Hma) complexed with peptidyl transferase substrate analogues that reveal an induced-fit mechanism in which substrates and active-site residues reposition to allow the peptidyl transferase reaction. Proper binding of an aminoacyl-tRNA analogue to the A site induces specific movements of 23S rRNA nucleotides 2618–2620 (Escherichia coli numbering 2583–2585) and 2541(2506), thereby reorienting the ester group of the peptidyl-tRNA and making it accessible for attack. In the absence of the appropriate A-site substrate, the peptidyl transferase centre positions the ester link of the peptidyl-tRNA in a conformation that precludes the catalysed nucleophilic attack by water. Protein release factors2 may also function, in part, by inducing an active-site rearrangement similar to that produced by the A-site aminoacyl-tRNA, allowing the carbonyl group and water to be positioned for hydrolysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Green, R. & Lorsch, J. R. The path to perdition is paved with protons. Cell 110, 665–668 (2002)
Kisselev, L. L. & Buckingham, R. H. Translational termination comes of age. Trends Biochem. Sci. 25, 561–566 (2000)
Koshland, D. E. in The Enzymes (eds Boyer, P. D., Lardy, H. & Myrback, K.) 305–346 (Academic, New York, 1959)
Bennett, W. S. Jr & Steitz, T. A. Glucose-induced conformational change in yeast hexokinase. Proc. Natl Acad. Sci. USA 75, 4848–4852 (1978)
Caskey, C. T., Beaudet, A. L., Scolnick, E. M. & Rosman, M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc. Natl Acad. Sci. USA 68, 3163–3167 (1971)
Zavialov, A. V., Mora, L., Buckingham, R. H. & Ehrenberg, M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10, 789–798 (2002)
Moazed, D. & Noller, H. F. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 (1989)
Pape, T., Wintermeyer, W. & Rodnina, M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18, 3800–3807 (1999)
Katunin, V. I., Muth, G. W., Strobel, S. A., Wintermeyer, W. & Rodnina, M. V. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell 10, 339–346 (2002)
Schmeing, T. M. et al. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nature Struct. Biol. 9, 225–230 (2002)
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000)
Agmon, I. et al. Ribosomal crystallography: a flexible nucleotide anchoring tRNA translocation, facilitates peptide-bond formation, chirality discrimination and antibiotics synergism. FEBS Lett. 567, 20–26 (2004)
Hansen, J. L., Schmeing, T. M., Moore, P. B. & Steitz, T. A. Structural insights into peptide bond formation. Proc. Natl Acad. Sci. USA 99, 11670–11675 (2002)
Burgi, H. B., Dunitz, J. D. & Shefter, E. Geometrical reaction coordinates. II. Nucleophilic addition to a carbonyl group. J. Am. Chem. Soc. 95, 5065–5067 (1973)
Radisky, E. S. & Koshland, D. E. Jr. A clogged gutter mechanism for protease inhibitors. Proc. Natl Acad. Sci. USA 99, 10316–10321 (2002)
Klein, D. J., Moore, P. B. & Steitz, T. A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340, 141–177 (2004)
Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B. & Steitz, T. A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA 98, 4899–4903 (2001)
Weinger, J. S., Parnell, K. M., Dorner, S., Green, R. & Strobel, S. A. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nature Struct. Mol. Biol. 11, 1101–1106 (2004)
Schmeing, T. M., Moore, P. B. & Steitz, T. A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9, 1345–1352 (2003)
Seila, A. C., Okuda, K., Nunez, S., Seila, A. F. & Strobel, S. A. Kinetic isotope effect analysis of the ribosomal peptidyl transferase reaction. Biochemistry 44, 4018–4027 (2005)
Scarlett, D. J., McCaughan, K. K., Wilson, D. N. & Tate, W. P. Mapping functionally important motifs SPF and GGQ of the decoding release factor RF2 to the Escherichia coli ribosome by hydroxyl radical footprinting. Implications for macromolecular mimicry and structural changes in RF2. J. Biol. Chem. 278, 15095–15104 (2003)
Rawat, U. B. et al. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421, 87–90 (2003)
Frolova, L. Y. et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020 (1999)
Youngman, E. M., Brunelle, J. L., Kochaniak, A. B. & Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 (2004)
Weinger, J. S., Kitchen, D., Scaringe, S. A., Strobel, S. A. & Muth, G. W. Solid phase synthesis and binding affinity of peptidyl transferase transition state mimics containing 2′-OH at P-site position A76. Nucleic Acids Res. 32, 1502–1511 (2004)
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905–920 (2000)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Acknowledgements
We thank E. Youngman and R. Green for allowing us to use their unpublished rates of peptidyl-tRNA hydrolysis; G. Muth, D. Kitchen and S. Scaringe for assistance with synthetic chemistry; J. Kavran for collection of a diffraction data set; A. Innis for reading the text; and P. Moore for experimental advice and critical reading of the manuscript. Financial support for the National Synchrotron Light Source comes from the US DOE and the NIH. The Advanced Light Source is supported by the DOE. K.S.H.. is supported by a NIH postdoctoral fellowship. This work was supported by NIH and ACS Beginning Investigator grants to S.A.S. and an NIH grant to T.A.S. Author Contributions T.M.S. determined and evaluated the structures of the three complexes of the 50S ribosomal subunit with substrate analogues under the supervision of T.A.S., and K.S.H. designed and synthesized the substrate analogues used under the supervision of S.A.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The atomic coordinates and structure factors have been deposited into the RCSB Protein Data Bank under accession codes 1VQ6, 1VQ7 and 1VQN. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
A table summarizing data and refinement statistics for structure determination. (DOC 52 kb)
Supplementary Figure 1
A schematic drawing of the peptidyl transferase reaction. (PDF 488 kb)
Supplementary Figure 2
Superimposition of structures of the large ribosomal subunit of H. marismortui in the uninduced state. (PDF 1857 kb)
Supplementary Figure 3
Superimposition of structures of the large ribosomal subunit of H. marismortui in the uninduced state. (PDF 1625 kb)
Supplementary Figure Legends
Text to accompany the Supplementary Figures. (DOC 20 kb)
Supplementary Video 1
A video clip demonstrating how proper binding of the A-site substrate induces changes to the conformation of the peptidyl transferase center and the P-site substrate. (MOV 3606 kb)
Supplementary Video 2
A close-up view of conformational changes associated with the induced fit mechanism. (MOV 10740 kb)
Supplementary Video Legends
Legends for the Supplementary Videos. (DOC 20 kb)
Rights and permissions
About this article
Cite this article
Martin Schmeing, T., Huang, K., Strobel, S. et al. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438, 520–524 (2005). https://doi.org/10.1038/nature04152
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04152
This article is cited by
-
Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it
Nature Chemical Biology (2024)
-
RAPP-containing arrest peptides induce translational stalling by short circuiting the ribosomal peptidyltransferase activity
Nature Communications (2024)
-
Insights into the ribosome function from the structures of non-arrested ribosome–nascent chain complexes
Nature Chemistry (2023)
-
Regulation of the macrolide resistance ABC-F translation factor MsrD
Nature Communications (2023)
-
Structural and mechanistic basis for translation inhibition by macrolide and ketolide antibiotics
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.