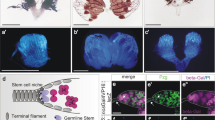
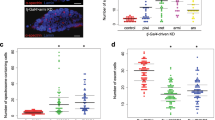
Abstract
Caenorhabditis elegans homologues of the retinoblastoma (Rb) tumour suppressor complex specify cell lineage during development1,2. Here we show that mutations in Rb pathway components enhance RNA interference (RNAi) and cause somatic cells to express genes and elaborate perinuclear structures normally limited to germline-specific P granules. Furthermore, particular gene inactivations that disrupt RNAi reverse the cell lineage transformations of Rb pathway mutants. These findings suggest that mutations in Rb pathway components cause cells to revert to patterns of gene expression normally restricted to germ cells. Rb may act by a similar mechanism to transform mammalian cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lu, X. & Horvitz, H. R. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95, 981–991 (1998)
Ceol, C. J. & Horvitz, H. R. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signalling in C. elegans vulval development. Mol. Cell 7, 461–473 (2001)
Huang, L. S., Tzou, P. & Sternberg, P. W. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5, 395–411 (1994)
Hsieh, J. et al. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13, 2958–2970 (1999)
Kennedy, S., Wang, D. & Ruvkun, G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645–649 (2004)
Simmer, F. et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317–1319 (2002)
Sieburth, D. et al. Systematic analysis of genes required for synapse structure and function. Nature doi:10.1038/nature03809 (this issue)
Sijen, T. et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 (2001)
Smardon, A. et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178 (2000)
Kawasaki, I. et al. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645 (1998)
Unhavaithaya, Y. et al. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111, 991–1002 (2002)
Pitt, J. N., Schisa, J. A. & Priess, J. R. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 219, 315–333 (2000)
Kim, J. K. et al. Functional genomic analysis of RNA interference in C. elegans. Science 308, 1164–1167 (2005)
Sijen, T. & Plasterk, R. H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426, 310–314 (2003)
Robert, V. J., Sijen, T., van Wolfswinkel, J. & Plasterk, R. H. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19, 782–787 (2005)
Klymenko, T. & Muller, J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 5, 373–377 (2004)
Debernardi, S. et al. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood 99, 275–281 (2002)
Linder, B., Gerlach, N. & Jackle, H. The Drosophila homolog of the human AF10 is an HP1-interacting suppressor of position effect variegation. EMBO Rep. 2, 211–216 (2001)
Braas, D., Gundelach, H. & Klempnauer, K. H. The glioma-amplified sequence 41 gene (GAS41) is a direct Myb target gene. Nucleic Acids Res. 32, 4750–4757 (2004)
Lai, A. et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumour suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 21, 2918–2932 (2001)
Lewis, P. W. et al. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 18, 2929–2940 (2004)
Fong, Y., Bender, L., Wang, W. & Strome, S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science 296, 2235–2238 (2002)
Myers, T. R. & Greenwald, I. lin-35 Rb acts in the major hypodermis to oppose ras-mediated vulval induction in C. elegans. Dev. Cell 8, 117–123 (2005)
Fischer, U., Meltzer, P. & Meese, E. Twelve amplified and expressed genes localized in a single domain in glioma. Hum. Genet. 98, 625–628 (1996)
Streubel, B. et al. Amplification of the MLL gene on double minutes, a homogeneously staining region, and ring chromosomes in five patients with acute myeloid leukemia or myelodysplastic syndrome. Genes Chromosom. Cancer 27, 380–386 (2000)
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004)
Pal-Bhadra, M. et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303, 669–672 (2004)
Couteau, F., Guerry, F., Muller, F. & Palladino, F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3, 235–241 (2002)
Black, E. P. et al. Distinct gene expression phenotypes of cells lacking Rb and Rb family members. Cancer Res. 63, 3716–3723 (2003)
Godbout, R., Packer, M. & Bie, W. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J. Biol. Chem. 273, 21161–21168 (1998)
Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. Stemness: transcriptional profiling of embryonic and adult stem cells. Science 298, 597–600 (2002)
Acknowledgements
We thank E. C. Andersen, W. G. Kelly, N. Dyson and V. Reineke for discussions; T. Sijen for reagents; H. Y. Mak for the unpublished tub-1::gfp transgene; J. Ahringer and M. Vidal for the feeding RNAi bacteria clones; S. Fischer for constructing the hand-picked RNAi library; S. Strome for the anti-PGL-1 antibodies; and E. C. Andersen, H. R. Horvitz, C. P. Hunter, F. Palladino, the Caenorhabditis Genetics Center and C. elegans Gene Knockout Consortium for some of the strains.Author Contributions D.W. conducted experiments and analysed data for all figures and tables, except for Figs 2a and 3b; S.K. made the initial observation of enhanced RNAi in lin-15 and synergistic enhancement of RNAi in eri-1; lin-15; D.C. did immunofluorescence staining of PGL-1 in Figs 1a and 3b; J.K.K., H.W.G. and R.S.K. identified 36 RNAi-defective genes that were tested by D.W. for their interaction with the Rb pathway; C.C.M. and G.R. were principal investigators.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
Additional descriptions of methods used in this study. (PDF 17 kb)
Supplemenetary Table S1
The enhanced RNAi, but not the cell lineage defects, of Rb pathway mutants requires the canonical RNAi pathway. (PDF 41 kb)
Supplementary Table S2
Mutation in lin-15B releases the requirement of RNAi for the RNA-dependent RNA polymerase RRF-1. (PDF 9 kb)
Rights and permissions
About this article
Cite this article
Wang, D., Kennedy, S., Conte, D. et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436, 593–597 (2005). https://doi.org/10.1038/nature04010
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04010
This article is cited by
-
G protein-coupled receptor-based thermosensation determines temperature acclimatization of Caenorhabditis elegans
Nature Communications (2024)
-
The head mesodermal cell couples FMRFamide neuropeptide signaling with rhythmic muscle contraction in C. elegans
Nature Communications (2023)
-
Mitochondrial hydrogen peroxide positively regulates neuropeptide secretion during diet-induced activation of the oxidative stress response
Nature Communications (2021)
-
Isolated C. elegans germ nuclei exhibit distinct genomic profiles of histone modification and gene expression
BMC Genomics (2019)
-
Somatically expressed germ-granule components, PGL-1 and PGL-3, repress programmed cell death in C. elegans
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.