Abstract
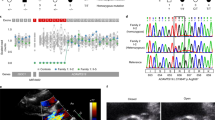
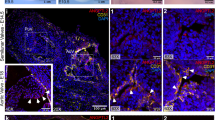
Calcification of the aortic valve is the third leading cause of heart disease in adults1. The incidence increases with age, and it is often associated with a bicuspid aortic valve present in 1–2% of the population2. Despite the frequency, neither the mechanisms of valve calcification nor the developmental origin of a two, rather than three, leaflet aortic valve is known. Here, we show that mutations in the signalling and transcriptional regulator NOTCH1 cause a spectrum of developmental aortic valve anomalies and severe valve calcification in non-syndromic autosomal-dominant human pedigrees. Consistent with the valve calcification phenotype, Notch1 transcripts were most abundant in the developing aortic valve of mice, and Notch1 repressed the activity of Runx2, a central transcriptional regulator of osteoblast cell fate. The hairy-related family of transcriptional repressors (Hrt), which are activated by Notch1 signalling, physically interacted with Runx2 and repressed Runx2 transcriptional activity independent of histone deacetylase activity. These results suggest that NOTCH1 mutations cause an early developmental defect in the aortic valve and a later de-repression of calcium deposition that causes progressive aortic valve disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
American Heart Association, Heart Disease and Stroke Statistics—2004 Update (American Heart Association, Dallas, Texas, 2004)
Hoffman, J. I. & Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 (2002)
Cripe, L., Andelfinger, G., Martin, L. J., Shooner, K. & Benson, D. W. Bicuspid aortic valve is heritable. J. Am. Coll. Cardiol. 44, 138–143 (2004)
Horne, B. D., Camp, N. J., Muhlestein, J. B. & Cannon-Albright, L. A. Evidence for a heritable component in death resulting from aortic and mitral valve diseases. Circulation 110, 3143–3148 (2004)
Loffredo, C. A. et al. Prevalence of congenital cardiovascular malformations among relatives of infants with hypoplastic left heart, coarctation of the aorta, and d-transposition of the great arteries. Am. J. Med. Genet. 124A, 225–230 (2004)
Rajamannan, N. M., Gersh, B. & Bonow, R. O. Calcific aortic stenosis: from bench to the bedside—emerging clinical and cellular concepts. Heart 89, 801–805 (2003)
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signalling: cell fate control and signal integration in development. Science 284, 770–776 (1999)
Frischmeyer, P. A. et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 (2002)
Krebs, L. T. et al. Notch signalling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343–1352 (2000)
Timmerman, L. A. et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18, 99–115 (2004)
Wen, C., Metzstein, M. M. & Greenwald, I. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development 124, 4759–4767 (1997)
Sotillos, S., Roch, F. & Campuzano, S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124, 4769–4779 (1997)
Struhl, G. & Adachi, A. Nuclear access and action of notch in vivo. Cell 93, 649–660 (1998)
Struhl, G. & Greenwald, I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398, 522–525 (1999)
Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997)
Haines, N. & Irvine, K. D. Glycosylation regulates Notch signalling. Nature Rev. Mol. Cell Biol. 4, 786–797 (2003)
O'Brien, K. D. et al. Osteopontin is expressed in human aortic valvular lesions. Circulation 92, 2163–2168 (1995)
Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L. & Karsenty, G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754 (1997)
Steitz, S. A. et al. Smooth muscle cell phenotypic transition associated with calcification: upregulation of Cbfa1 and downregulation of smooth muscle lineage markers. Circ. Res. 89, 1147–1154 (2001)
Rajamannan, N. M. et al. Atorvastatin inhibits hypercholesterolemia-induced cellular proliferation and bone matrix production in the rabbit aortic valve. Circulation 105, 2660–2665 (2002)
Vega, R. B. et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555–566 (2004)
Nakagawa, O., Nakagawa, M., Richardson, J. A., Olson, E. N. & Srivastava, D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev. Biol. 216, 72–84 (1999)
Nakagawa, O. et al. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signalling. Proc. Natl Acad. Sci. USA 97, 13655–13660 (2000)
McLarren, K. W. et al. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J. Biol. Chem. 275, 530–538 (2000)
Iso, T. et al. HERP, a novel heterodimer partner of HES/E(spl) in Notch signalling. Mol. Cell. Biol. 21, 6080–6089 (2001)
Weng, A. P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004)
Garg, V. et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447 (2003)
Kruglyak, L., Daly, M. J., Reeve-Daly, M. P. & Lander, E. S. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 58, 1347–1363 (1996)
Cohen, J. et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nature Genet. 37, 161–165 (2005)
Kathiriya, I. S. et al. Hairy-related transcription factors inhibit GATA-dependent cardiac gene expression through a signal-responsive mechanism. J. Biol. Chem. 279, 54937–54943 (2004)
Acknowledgements
The authors thank the families for their participation; the Divisions of Pediatric Cardiology and Pediatric Cardiothoracic Surgery at Children's Medical Center Dallas for assistance with clinical information; McDermott Center for Human Growth and Development for assistance with linkage analysis and allelic discrimination assays; Dallas Heart Study participants and investigators for DNA samples; members of the Molecular Histology Core laboratory for radioactive-section in situ hybridization; J. C. Cohen and H. H. Hobbs for discussions and critical review of this manuscript; K. Ivey for graphics assistance; C. Butler, A. Garg and D. Srivastava for assistance with blood collection; G. Karsenty for Runx2 expression and p6OSE2 reporter plasmids; and L. Kedes for the GST–Hrt2 expression plasmid. This work was supported by grants from NICHD/NIH and March of Dimes Birth Defects Foundation to V.G., and NHLBI/NIH, March of Dimes Birth Defects Foundation and the Donald W. Reynolds Cardiovascular Clinical Research Center to D.S. J.F.R. was supported by a training grant from NIH; I.N.K. is an NICHD/NIH fellow of the Pediatric Scientist Development Program; and D.S. is an Established Investigator of the American Heart Association.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure Legends
Text to accompany the below Supplementary Figures. (DOC 20 kb)
Supplementary Figure S1
Clinical phenotype of Family A and Family B. (PDF 1217 kb)
Supplementary Figure S2
Pedigree of Family A with haplotype data. (PDF 155 kb)
Supplementary Figure S3
Ethnicity data for NOTCH1 polymorphisms. (PDF 111 kb)
Rights and permissions
About this article
Cite this article
Garg, V., Muth, A., Ransom, J. et al. Mutations in NOTCH1 cause aortic valve disease. Nature 437, 270–274 (2005). https://doi.org/10.1038/nature03940
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature03940
This article is cited by
-
Enhancing aortic valve drug delivery with PAR2-targeting magnetic nano-cargoes for calcification alleviation
Nature Communications (2024)
-
Calcific aortic valve disease: mechanisms, prevention and treatment
Nature Reviews Cardiology (2023)
-
Left ventricular hypertrophy and metabolic resetting in the Notch3-deficient adult mouse heart
Scientific Reports (2023)
-
Multiscale computational modeling of aortic valve calcification
Biomechanics and Modeling in Mechanobiology (2023)
-
The Advent of Spatial Omics in Congenital Heart Disease
Current Treatment Options in Pediatrics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.