Abstract
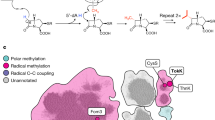
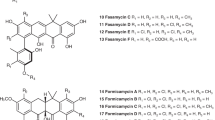
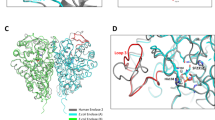
The biosynthetic pathway of the clinically important antibiotic fosfomycin uses enzymes that catalyse reactions without precedent in biology. Among these is hydroxypropylphosphonic acid epoxidase, which represents a new subfamily of non-haem mononuclear iron enzymes. Here we present six X-ray structures of this enzyme: the apoenzyme at 2.0 Å resolution; a native Fe(ii)-bound form at 2.4 Å resolution; a tris(hydroxymethyl)aminomethane–Co(ii)-enzyme complex structure at 1.8 Å resolution; a substrate–Co(ii)-enzyme complex structure at 2.5 Å resolution; and two substrate–Fe(ii)-enzyme complexes at 2.1 and 2.3 Å resolution. These structural data lead us to suggest how this enzyme is able to recognize and respond to its substrate with a conformational change that protects the radical-based intermediates formed during catalysis. Comparisons with other family members suggest why substrate binding is able to prime iron for dioxygen binding in the absence of α-ketoglutarate (a co-substrate required by many mononuclear iron enzymes), and how the unique epoxidation reaction of hydroxypropylphosphonic acid epoxidase may occur.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Liu, P. et al. Protein purification and functional assignment of the epoxidase catalyzing the formation of fosfomycin. J. Am. Chem. Soc. 123, 4619–4620 (2001)
Lobel, B. Short term therapy for uncomplicated urinary tract infection today. Clinical outcome upholds the theories. Int. J. Antimicrob. Agents 22, 85–87 (2003)
Nakazawa, H., Kikuchi, Y., Honda, T., Isago, T. & Nozaki, M. Enhancement of antimicrobial effects of various antibiotics against methicillin-resistant Staphylococcus aureus (MRSA) by combination with fosfomycin. J. Infect. Chemother. 9, 304–309 (2003)
Cassone, M., Campanile, F., Pantosti, A., Venditti, M. & Stefani, S. Identification of a variant “Rome clone” of methicillin-resistant Staphylococcus aureus with decreased susceptibility to vancomycin, responsible for an outbreak in an intensive care unit. Microb. Drug Resist. 10, 43–49 (2004)
Hidaka, T. et al. Cloning and nucleotide sequence of fosfomycin biosynthetic genes of Streptomyces wedmorensis. Mol. Gen. Genet. 249, 274–280 (1995)
Hammerschmidt, F. Biosynthesis of natural-products with a P-C bond. 8. On the origin of the oxirane oxygen atom of fosfomycin in Streptomyces fradiae. J. Chem. Soc. Perkin Trans. 1 8, 1993–1996 (1991)
Liu, P. et al. Biochemical and spectroscopic studies on (S)-2-hydroxypropylphosphonic acid epoxidase: a novel mononuclear non-heme iron enzyme. Biochemistry 42, 11577–11586 (2003)
Guengerich, F. P. Cytochrome P450 oxidations in the generation of reactive electrophiles: epoxidation and related reactions. Arch. Biochem. Biophys. 409, 59–71 (2003)
Costas, M., Mehn, M. P., Jensen, M. P. & Que, L. Jr Dioxygen activation at mononuclear nonheme iron active sites: enzymes, models, and intermediates. Chem. Rev. 104, 939–986 (2004)
Dunwell, J. M. Cupins: a new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol. Genet. Eng. Rev. 15, 1–32 (1998)
Zhao, Z. et al. Mechanistic studies of HPP epoxidase: configuration of the substrate governs its enzymatic fate. Angew. Chem. Int. Edn Engl. 41, 4529–4532 (2002)
Roach, P. L. et al. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 387, 827–830 (1997)
Dunwell, J. M., Culham, A., Carter, C. E., Sosa-Aguirre, C. R. & Goodenough, P. W. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26, 740–746 (2001)
Dunwell, J. M., Purvis, A. & Khuri, S. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65, 7–17 (2004)
Hausinger, R. P. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 21–68 (2004)
Liu, P. et al. Oxygenase activity in the self-hydroxylation of (S)-2-hydroxypropylphosphonic acid epoxidase involved in fosfomycin biosynthesis. J. Am. Chem. Soc. 126, 10306–10312 (2004)
Ryle, M. J. et al. O2- and α-ketoglutarate-dependent tyrosyl radical formation in TauD, an α-keto acid-dependent non-heme iron dioxygenase. Biochemistry 42, 1854–1862 (2003)
Roach, P. L., Schofield, C. J., Baldwin, J. E., Clifton, I. J. & Hajdu, J. Crystallization and preliminary X-ray diffraction studies on recombinant isopenicillin N synthase from Aspergillus nidulans. Protein Sci. 4, 1007–1009 (1995)
Valegård, K. et al. The structural basis of cephalosporin formation in a mononuclear ferrous enzyme. Nature Struct. Mol. Biol. 11, 95–101 (2004)
Price, J. C., Barr, E. W., Tirupati, B., Bollinger, J. M. Jr & Krebs, C. The first direct characterization of a high-valent iron intermediate in the reaction of an α-ketoglutarate-dependent dioxygenase: a high-spin FeIV complex in taurine/α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry 42, 7497–7508 (2003)
Zhou, J. et al. Spectroscopic studies of substrate interactions with clavaminate synthase 2, a multifunctional α-KG-dependent non-heme iron enzyme: correlation with mechanisms and reactivities. J. Am. Chem. Soc. 123, 7388–7398 (2001)
Solomon, E. I. et al. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem. Rev. 100, 235–350 (2000)
Roach, P. L. et al. Anaerobic crystallisation of an isopenicillin N synthase.Fe(II).substrate complex demonstrated by X-ray studies. Eur. J. Biochem. 242, 736–740 (1996)
Hegg, E. L. & Que, L. Jr The 2-His-1-carboxylate facial triad—an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 250, 625–629 (1997)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Terwilliger, T. C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D 59, 38–44 (2003)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Navaza, J. AMoRe: an Automated Package for Molecular Replacement. Acta Crystallogr. A50, 164–182 (1994)
Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D 55, 484–491 (1999)
Laskowski, R., MacArthur, M., Moss, D. & Thornton, J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, California, 2002)
Acknowledgements
This research is supported in part by the National Institutes of Health (C.L.D. and H-W.L.), the National Institute of Environmental Health Sciences (L.J.H.), the Searle Scholars Program (C.L.D.), Alfred P. Sloan Foundation (C.L.D.), and a Lester Wolfe Predoctoral Fellowship (L.J.H.). Synchrotron facilities are funded by the NIH National Center of Research Resources (Advanced Photon Source NE-CAT 8BM) and the US Department of Energy, Division of Materials Sciences and Division of Chemical Sciences (National Synchrotron Light Source, Brookhaven National Laboratory).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Atomic coordinates for the following structures have been deposited in the Protein Data Bank: apo-HppE (1ZZ6), Fe(ii)-HppE (1ZZ9), form-1 S-HPP–Fe(ii)-HppE (1ZZ7), form-2 S-HPP–Fe(ii)-HppE (1ZZ8), Tris–Co(ii)-HppE (1ZZC) and S-HPP–Co(ii)-HppE (1ZZB). Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
Site-directed mutagenesis, protein purification, assays, crystallization and data collection.
Supplementary Figure S1
Stereoviews of HppE active sites displayed with 2Fo-Fc maps contoured from 1.0-1.5ω .
Supplementary Figure S2
Structural insights into catalysis (stereoviews).
Supplementary Figure S3
Stereoviews of S-HPP-Co(II)-HppE active sites displayed with 2Fo-Fc maps contoured from 1.0-1.5ω.
Supplementary Tables
Supplementary Table S1 details data and refinement statistics for Co and Apo HppE structures. Supplementary Table S2 details data and refinement statistics for Fe HppE structures
Rights and permissions
About this article
Cite this article
Higgins, L., Yan, F., Liu, P. et al. Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme. Nature 437, 838–844 (2005). https://doi.org/10.1038/nature03924
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature03924
This article is cited by
-
Engineering non-haem iron enzymes for enantioselective C(sp3)–F bond formation via radical fluorine transfer
Nature Synthesis (2024)
-
Advances in Fe(II)/2-ketoglutarate-dependent dioxygenase-mediated C–H bond oxidation for regioselective and stereoselective hydroxyl amino acid synthesis: from structural insights into practical applications
Systems Microbiology and Biomanufacturing (2021)
-
Dioxygen activation by nonheme iron enzymes with the 2-His-1-carboxylate facial triad that generate high-valent oxoiron oxidants
JBIC Journal of Biological Inorganic Chemistry (2017)
-
Go it alone: four-electron oxidations by mononuclear non-heme iron enzymes
JBIC Journal of Biological Inorganic Chemistry (2017)
-
Beyond ferryl-mediated hydroxylation: 40 years of the rebound mechanism and C–H activation
JBIC Journal of Biological Inorganic Chemistry (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.