Abstract
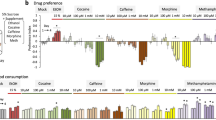
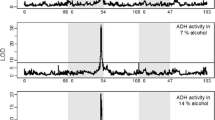
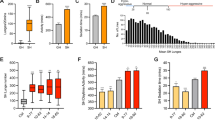
Repeated alcohol consumption leads to the development of tolerance, simply defined as an acquired resistance to the physiological and behavioural effects of the drug. This tolerance allows increased alcohol consumption, which over time leads to physical dependence and possibly addiction1,2,3. Previous studies have shown that Drosophila develop ethanol tolerance, with kinetics of acquisition and dissipation that mimic those seen in mammals. This tolerance requires the catecholamine octopamine, the functional analogue of mammalian noradrenaline4. Here we describe a new gene, hangover, which is required for normal development of ethanol tolerance. hangover flies are also defective in responses to environmental stressors, such as heat and the free-radical-generating agent paraquat. Using genetic epistasis tests, we show that ethanol tolerance in Drosophila relies on two distinct molecular pathways: a cellular stress pathway defined by hangover, and a parallel pathway requiring octopamine. hangover encodes a large nuclear zinc-finger protein, suggesting a role in nucleic acid binding. There is growing recognition that stress, at both the cellular and systemic levels, contributes to drug- and addiction-related behaviours in mammals. Our studies suggest that this role may be conserved across evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tabakoff, B., Cornell, N. & Hoffman, P. L. Alcohol tolerance. Ann. Emerg. Med. 15, 1005–1012 (1986)
Lê, A. D. & Mayer, J. M. in Pharmacological Effects of Ethanol on the Nervous System (eds Deitrich, R. A. & Erwin, V. G.) 251–268 (CRC Press, Boca Raton, 1996)
Fadda, F. & Rossetti, Z. L. Chronic ethanol consumption: from neuroadaptation to neurodegeneration. Prog. Neurobiol. 56, 385–431 (1998)
Scholz, H., Ramond, J., Singh, C. M. & Heberlein, U. Functional ethanol tolerance in Drosophila. Neuron 28, 261–271 (2000)
Weber, K. E. & Diggins, L. T. Increased selection response in larger populations. II. Selection for ethanol vapor resistance in Drosophila melanogaster at two population sizes. Genetics 125, 585–597 (1990)
Moore, M. S. et al. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signalling pathway. Cell 93, 997–1007 (1998)
Singh, C. M. & Heberlein, U. Genetic control of acute ethanol-induced behaviors in Drosophila. Alcohol. Clin. Exp. Res. 24, 1127–1136 (2000)
Legrain, P. & Choulika, A. The molecular characterization of PRP6 and PRP9 yeast genes reveals a new cysteine/histidine motif common to several splicing factors. EMBO J. 9, 2775–2781 (1990)
Torroja, L., Chu, H., Kotovsky, I. & White, K. Neuronal overexpression of APPL, the Drosophila homologue of the amyloid precursor protein (APP), disrupts axonal transport. Curr. Biol. 9, 489–492 (1999)
Wilke, N., Sganga, M., Barhite, S. & Miles, M. F. Effects of alcohol on gene expression in neural cells. EXS 71, 49–59 (1994)
Piper, P. W. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 134, 121–127 (1995)
Monastirioti, M., Linn, C. E. J. & White, K. Characterization of Drosophila tyramine β-hydroxylase gene and isolation of mutant flies lacking octopamine. J. Neurosci. 16, 3900–3911 (1996)
Arking, R., Buck, S., Berrios, A., Dwyer, S. & Baker, G. T. III Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev. Genet. 12, 362–370 (1991)
Finkel, T. & Holbrook, N. J. Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247 (2000)
Min, K. T. & Benzer, S. Spongecake and eggroll: two hereditary diseases in Drosophila resemble patterns of human brain degeneration. Curr. Biol. 7, 885–888 (1997)
Suzuki, Y. J., Forman, H. J. & Sevanian, A. Oxidants as stimulators of signal transduction. Free Radic. Biol. Med. 22, 269–285 (1997)
Walter, H. J. & Messing, R. O. Regulation of neuronal voltage-gated calcium channels by ethanol. Neurochem. Int. 35, 95–101 (1999)
Sun, A. Y. et al. Ethanol and oxidative stress. Alcohol. Clin. Exp. Res. 25, 237S–243S (2001)
Schwaerzel, M. et al. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J. Neurosci. 23, 10495–10502 (2003)
Hyman, S. E. & Malenka, R. C. Addiction and the brain: the neurobiology of compulsion and its persistence. Nature Rev. Neurosci. 2, 695–703 (2001)
Nestler, E. J. Molecular basis of long-term plasticity underlying addiction. Nature Rev. Neurosci. 2, 119–128 (2001)
Mlodzik, M. & Hiromi, Y. Enhancer trap method in Drosophila: Its application to neurobiology. Methods Neurosci. 9, 397–414 (1992)
Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993)
Acknowledgements
We thank R. Threlkeld for the northern blot, N. Funk and I. Schwenkert for the production of rescue constructs and anti-HANG antibody, H.Saumweber and G.Krohne for gifts of antibodies, and B. Poeck, A. Corl, A. Rothenfluh, D. Guarnieri, F. Wolf and C. Kenyon for comments on the manuscript. This work was supported by the NIH/NIAAA (U.H.), ABMRF (U.H.), the Sandler Foundation (U.H.), the Wheeler Center (H.S. and U.H.) and the German Science Foundation DFG (H.S.)
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure S1
a, hangAE10 flies show normal ethanol absorption and metabolism. b, The hangAE10 phenotype is reverted to wild type after excision of the P-element. (JPG 201 kb)
Supplementary Figure 2
hang is expressed ubiquitously in the nervous system. (JPG 459 kb)
Supplementary Figure S3
Hang flies show reduced lifespan but no obvious neurodegeneration. (JPG 406 kb)
Rights and permissions
About this article
Cite this article
Scholz, H., Franz, M. & Heberlein, U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature 436, 845–847 (2005). https://doi.org/10.1038/nature03864
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03864
This article is cited by
-
Aedes aegypti miRNA-33 modulates permethrin induced toxicity by regulating VGSC transcripts
Scientific Reports (2021)
-
Studying alcohol use disorder using Drosophila melanogaster in the era of ‘Big Data’
Behavioral and Brain Functions (2019)
-
Genetic analysis of variation in lifespan using a multiparental advanced intercross Drosophila mapping population
BMC Genetics (2016)
-
Cross-stress resistance in Saccharomyces cerevisiae yeast—new insight into an old phenomenon
Cell Stress and Chaperones (2016)
-
Polymorphisms in early neurodevelopmental genes affect natural variation in alcohol sensitivity in adult drosophila
BMC Genomics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.