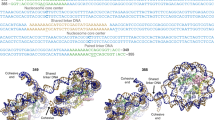
Abstract
DNA in eukaryotic chromosomes is organized in arrays of nucleosomes compacted into chromatin fibres. This higher-order structure of nucleosomes is the substrate for DNA replication, recombination, transcription and repair. Although the structure of the nucleosome core is known at near-atomic resolution1, even the most fundamental information about the organization of nucleosomes in the fibre is controversial. Here we report the crystal structure of an oligonucleosome (a compact tetranucleosome) at 9 Å resolution, solved by molecular replacement using the nucleosome core structure. The structure shows that linker DNA zigzags back and forth between two stacks of nucleosome cores, which form a truncated two-start helix, and does not follow a path compatible with a one-start solenoidal helix2. The length of linker DNA is most probably buffered by stretching of the DNA contained in the nucleosome cores. We have built continuous fibre models by successively stacking tetranucleosomes one on another. The resulting models are nearly fully compacted and most closely resemble the previously described crossed-linker model3. They suggest that the interfaces between nucleosomes along a single helix start are polymorphic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Davey, C. A., Sargent, D. F., Luger, K., Maeder, A. W. & Richmond, T. J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319, 1097–1113 (2002)
Finch, J. T. & Klug, A. Solenoidal model for superstructure in chromatin. Proc. Natl Acad. Sci. USA 73, 1897–1901 (1976)
Williams, S. P. et al. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys. J. 49, 233–248 (1986)
Richmond, T. J. & Davey, C. A. The structure of DNA in the nucleosome core. Nature 423, 145–150 (2003)
Oudet, P., Gross-Bellard, M. & Chambon, P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4, 281–300 (1975)
Thoma, F. & Koller, T. Influence of histone H1 on chromatin structure. Cell 12, 101–107 (1977)
Ris, H. & Korenberg, J. in Cell Biology (eds Prescott, D. M. & Goldstein, L.) 268–361 (Academic, New York, 1979)
Woodcock, C. L., Frado, L. L. & Rattner, J. B. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J. Cell Biol. 99, 42–52 (1984)
Widom, J., Finch, J. T. & Thomas, J. O. Higher-order structure of long repeat chromatin. EMBO J. 4, 3189–3194 (1985)
Woodcock, C. L. Chromatin fibers observed in situ in frozen hydrated sections. Native fiber diameter is not correlated with nucleosome repeat length. J. Cell Biol. 125, 11–19 (1994)
Butler, P. J. & Thomas, J. O. Changes in chromatin folding in solution. J. Mol. Biol. 140, 505–529 (1980)
McGhee, J. D., Nickol, J. M., Felsenfeld, G. & Rau, D. C. Higher order structure of chromatin: Orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell 33, 831–841 (1983)
Athey, B. D., Smith, M. F., Rankert, D. A., Williams, S. P. & Langmore, J. P. The diameters of frozen-hydrated chromatin fibers increase with DNA linker length: evidence in support of variable diameter models for chromatin. J. Cell Biol. 111, 795–806 (1990)
Horowitz, R. A., Agard, D. A., Sedat, J. W. & Woodcock, C. L. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J. Cell Biol. 125, 1–10 (1994)
Pehrson, J. R. Thymine dimer formation as a probe of the path of DNA in and between nucleosomes in intact chromatin. Proc. Natl Acad. Sci. USA 86, 9149–9153 (1989)
Dorigo, B., Schalch, T., Bystricky, K. & Richmond, T. J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003)
Dorigo, B. et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306, 1571–1573 (2004)
Thomas, J. O. & Furber, V. Yeast chromatin structure. FEBS Lett. 66, 274–280 (1976)
Allan, J., Rau, D. C., Harborne, N. & Gould, H. Higher order structure in a short repeat length chromatin. J. Cell Biol. 98, 1320–1327 (1984)
Luger, K., Rechsteiner, T. J., Flaus, A. J., Waye, M. M. & Richmond, T. J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301–311 (1997)
Gerchman, S. E. & Ramakrishnan, V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc. Natl Acad. Sci. USA 84, 7802–7806 (1987)
Sen, D., Mitra, S. & Crothers, D. M. Higher order structure of chromatin: evidence from photochemically detected linear dichroism. Biochemistry 25, 3441–3447 (1986)
Widom, J. & Klug, A. Structure of the 300 Å chromatin filament: X-ray diffraction from oriented samples. Cell 43, 207–213 (1985)
Graziano, V., Gerchman, S. E., Schneider, D. K. & Ramakrishnan, V. Histone H1 is located in the interior of the chromatin 30-nm filament. Nature 368, 351–354 (1994)
Blank, T. A. & Becker, P. B. Electrostatic mechanism of nucleosome spacing. J. Mol. Biol. 252, 305–313 (1995)
Korber, P. & Horz, W. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J. Biol. Chem. 279, 35113–35120 (2004)
Read, R. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D 57, 1373–1382 (2001)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Acknowledgements
We thank C. Schulze-Briese, T. Tomizaki and A. Wagner for assistance in data collection, and J. Widom for providing us with the 601 sequence.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Coordinates have been deposited in the Protein Data Bank with identifier 1zbb. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure S1
Explanation of the packing disorder found in the tetranucleosome crystals. (PDF 96 kb)
Supplementary Figure S2
Small angle X-ray scattering (SAXS) distributions for chicken erythrocyte chromatin and three chromatin fibre models. (PDF 54 kb)
Supplementary Methods
Description of the sequence and preparation of the DNA contained in the tetranucleosome. (PDF 11 kb)
Supplementary Table S1
Statistics of the X-ray crystallographic structure determination. (PDF 50 kb)
Supplementary Video S1
Tetranucleosome with axis system displayed (video version of Fig. 1c). (WMV 7692 kb)
Rights and permissions
About this article
Cite this article
Schalch, T., Duda, S., Sargent, D. et al. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436, 138–141 (2005). https://doi.org/10.1038/nature03686
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03686
This article is cited by
-
Structure of the ISW1a complex bound to the dinucleosome
Nature Structural & Molecular Biology (2024)
-
Histone variant H2A.Z modulates nucleosome dynamics to promote DNA accessibility
Nature Communications (2023)
-
CENP-A and CENP-B collaborate to create an open centromeric chromatin state
Nature Communications (2023)
-
Cooperation between bHLH transcription factors and histones for DNA access
Nature (2023)
-
Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.