Abstract
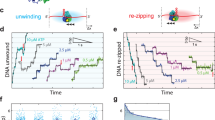
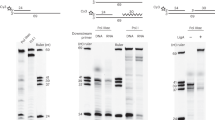
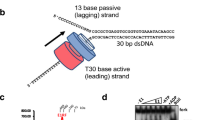
Helicases are molecular motors that use the energy of nucleoside 5′-triphosphate (NTP) hydrolysis to translocate along a nucleic acid strand and catalyse reactions such as DNA unwinding. The ring-shaped helicase1 of bacteriophage T7 translocates along single-stranded (ss)DNA at a speed of 130 bases per second2; however, T7 helicase slows down nearly tenfold when unwinding the strands of duplex DNA3. Here, we report that T7 DNA polymerase, which is unable to catalyse strand displacement DNA synthesis by itself, can increase the unwinding rate to 114 base pairs per second, bringing the helicase up to similar speeds compared to its translocation along ssDNA. The helicase rate of stimulation depends upon the DNA synthesis rate and does not rely on specific interactions between T7 DNA polymerase and the carboxy-terminal residues of T7 helicase. Efficient duplex DNA synthesis is achieved only by the combined action of the helicase and polymerase. The strand displacement DNA synthesis by the DNA polymerase depends on the unwinding activity of the helicase, which provides ssDNA template. The rapid trapping of the ssDNA bases by the DNA synthesis activity of the polymerase in turn drives the helicase to move forward through duplex DNA at speeds similar to those observed along ssDNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Patel, S. S. & Picha, K. M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 (2000)
Kim, D. E., Narayan, M. & Patel, S. S. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol. 321, 807–819 (2002)
Jeong, Y. J., Levin, M. K. & Patel, S. S. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl Acad. Sci. USA 101, 7264–7269 (2004)
Richardson, C. C. Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell 33, 315–317 (1983)
Lee, J., Chastain, P. D., Kusakabe, T., Griffith, J. D. & Richardson, C. C. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell 1, 1001–1010 (1998)
Patel, S. S., Wong, I. & Johnson, K. A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry 30, 511–525 (1991)
Matson, S. W. & Richardson, C. C. DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J. Biol. Chem. 258, 14009–14016 (1983)
Matson, S. W., Tabor, S. & Richardson, C. C. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 258, 14017–14024 (1983)
Patel, S. S., Rosenberg, A. H., Studier, F. W. & Johnson, K. A. Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J. Biol. Chem. 267, 15013–15021 (1992)
Ali, J. A. & Lohman, T. M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275, 377–380 (1997)
Notarnicola, S. M., Mulcahy, H. L., Lee, J. & Richardson, C. C. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J. Biol. Chem. 272, 18425–18433 (1997)
von Hippel, P. H. & Delagoutte, E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell 104, 177–190 (2001)
Kim, Y. T., Tabor, S., Bortner, C., Griffith, J. D. & Richardson, C. C. Purification and characterization of the bacteriophage T7 gene 2.5 protein. A single-stranded DNA-binding protein. J. Biol. Chem. 267, 15022–15031 (1992)
Lohman, T. M. & Ferrari, M. E. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63, 527–570 (1994)
Delagoutte, E. & von Hippel, P. H. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry 40, 4459–4477 (2001)
Levin, M. K. & Patel, S. S. in Molecular Motors (ed. Schliwa, M.) 179–198 (Wiley, Weinheim, 2003)
Levin, M. K., Gurjar, M. M. & Patel, S. S. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 278, 23311–23316 (2003)
Wang, H. & Oster, G. Ratchets, power strokes, and molecular motors. Applied Phys. A 75, 315–323 (2002)
Kim, S., Dallmann, H. G., McHenry, C. S. & Marians, K. J. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell 84, 643–650 (1996)
Mok, M. & Marians, K. J. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J. Biol. Chem. 262, 16644–16654 (1987)
Korhonen, J. A., Pham, X. H., Pellegrini, M. & Falkenberg, M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 (2004)
Schrock, R. D. & Alberts, B. Processivity of the gene 41 DNA helicase at the bacteriophage T4 DNA replication fork. J. Biol. Chem. 271, 16678–16682 (1996)
Cha, T. A. & Alberts, B. M. The bacteriophage T4 DNA replication fork. Only DNA helicase is required for leading strand DNA synthesis by the DNA polymerase holoenzyme. J. Biol. Chem. 264, 12220–12225 (1989)
Frey, M. W., Nossal, N. G., Capson, T. L. & Benkovic, S. J. Construction and characterization of a bacteriophage T4 DNA polymerase deficient in 3′ → 5′ exonuclease activity. Proc. Natl Acad. Sci. USA 90, 2579–2583 (1993)
Lucius, A. L., Maluf, N. K., Fischer, C. J. & Lohman, T. M. General methods for analysis of sequential “n-step” kinetic mechanisms: Application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85, 2224–2239 (2003)
Acknowledgements
We thank M. O'Donnell, A. Berdis, N. Andraos, C. C. Richardson and M. Salas for the gift of proteins, and C. M. Drain for critical reading of the manuscript. This research was supported by an NIH grant to S.S.P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
The effect of ssDNA-binding proteins on the unwinding activity of T7 helicase. (JPG 19 kb)
Supplementary Legend
This file contains the legend for Supplementary Fig. 1. (DOC 21 kb)
Rights and permissions
About this article
Cite this article
Stano, N., Jeong, YJ., Donmez, I. et al. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 435, 370–373 (2005). https://doi.org/10.1038/nature03615
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03615
This article is cited by
-
Continuous millisecond conformational cycle of a DEAH box helicase reveals control of domain motions by atomic-scale transitions
Communications Biology (2023)
-
Rad53 limits CMG helicase uncoupling from DNA synthesis at replication forks
Nature Structural & Molecular Biology (2020)
-
Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase
Nature Communications (2020)
-
The mechanism of DNA unwinding by the eukaryotic replicative helicase
Nature Communications (2019)
-
Helicase promotes replication re-initiation from an RNA transcript
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.