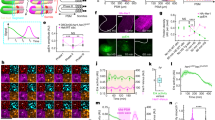
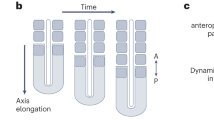
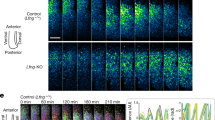
Abstract
The serially segmented (metameric) structures of vertebrates are based on somites that are periodically formed during embryogenesis. A ‘clock and wavefront’ model has been proposed to explain the underlying mechanism of somite formation1, in which the periodicity is generated by oscillation of Notch components (the clock) in the posterior pre-somitic mesoderm (PSM)2,3,4,5,6. This temporal periodicity is then translated into the segmental units in the ‘wavefront’7,8. The wavefront is thought to exist in the anterior PSM and progress backwards at a constant rate; however, there has been no direct evidence as to whether the levels of Notch activity really oscillate and how such oscillation is translated into a segmental pattern in the anterior PSM. Here, we have visualized endogenous levels of Notch1 activity in mice, showing that it oscillates in the posterior PSM but is arrested in the anterior PSM. Somite boundaries formed at the interface between Notch1-activated and -repressed domains. Genetic and biochemical studies indicate that this interface is generated by suppression of Notch activity by mesoderm posterior 2 (Mesp2) through induction of the lunatic fringe gene (Lfng). We propose that the oscillation of Notch activity is arrested and translated in the wavefront by Mesp2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cooke, J. & Zeeman, E. C. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 58, 455–476 (1976)
Palmeirim, I., Henrique, D., Ish-Horowicz, D. & Pourquie, O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648 (1997)
McGrew, M. J., Dale, J. K., Fraboulet, S. & Pourquie, O. The lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol. 8, 979–982 (1998)
Forsberg, H., Crozet, F. & Brown, N. A. Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr. Biol. 8, 1027–1030 (1998)
Jiang, Y. J. et al. Notch signalling and the synchronization of the somite segmentation clock. Nature 408, 475–479 (2000)
Bessho, Y. et al. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647 (2001)
Saga, Y. & Takeda, H. The making of the somite: molecular events in vertebrate segmentation. Nature Rev. Genet. 2, 835–845 (2001)
Pourquie, O. The segmentation clock: converting embryonic time into spatial pattern. Science 301, 328–330 (2003)
Saga, Y., Hata, N., Koseki, H. & Taketo, M. M. Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev. 11, 1827–1839 (1997)
Buchberger, A., Seidl, K., Klein, C., Eberhardt, H. & Arnold, H. H. cMeso-1, a novel bHLH transcription factor, is involved in somite formation in chicken embryos. Dev. Biol. 199, 201–215 (1998)
Sparrow, D. B. et al. Thylacine 1 is expressed segmentally within the paraxial mesoderm of the Xenopus embryo and interacts with the Notch pathway. Development 125, 2041–2051 (1998)
Sawada, A. et al. Zebrafish Mesp family genes, mesp-a and mesp-b are segmentally expressed in the presomitic mesoderm, and Mesp-b confers the anterior identity to the developing somites. Development 127, 1691–1702 (2000)
Takahashi, Y. et al. Mesp2 initiates somite segmentation through the Notch signalling pathway. Nature Genet. 25, 390–396 (2000)
Bruckner, K., Perez, L., Clausen, H. & Cohen, S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406, 411–415 (2000)
Moloney, D. J. et al. Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 (2000)
Dale, J. K. et al. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature 421, 275–278 (2003)
Bessho, Y., Hirata, H., Masamizu, Y. & Kageyama, R. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456 (2003)
Hrabe de Angelis, M., McIntyre, J. II & Gossler, A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature 386, 717–721 (1997)
Evrard, Y. A., Lun, Y., Aulehla, A., Gan, L. & Johnson, R. L. lunatic fringe is an essential mediator of somite segmentation and patterning. Nature 394, 377–381 (1998)
Cole, S. E., Levorse, J. M., Tilghman, S. M. & Vogt, T. F. Clock regulatory elements control cyclic expression of Lunatic fringe during somitogenesis. Dev. Cell 3, 75–84 (2002)
Morales, A. V., Yasuda, Y. & Ish-Horowicz, D. Periodic Lunatic fringe expression is controlled during segmentation by a cyclic transcriptional enhancer responsive to notch signaling. Dev. Cell 3, 63–74 (2002)
Bessho, Y. et al. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647 (2001)
Sawada, A. et al. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128, 4873–4880 (2001)
Dubrulle, J., McGrew, M. J. & Pourquie, O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell 106, 219–232 (2001)
Panin, V. M., Papayannopoulos, V., Wilson, R. & Irvine, K. D. Fringe modulates Notch–ligand interactions. Nature 387, 908–912 (1997)
Haines, N. & Irvine, K. D. Glycosylation regulates Notch signalling. Nature Rev. Mol. Cell Biol. 4, 786–797 (2003)
Hicks, C. et al. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nature Cell Biol. 2, 515–520 (2000)
Moreno, T. A. & Kintner, C. Regulation of segmental patterning by retinoic acid signaling during Xenopus somitogenesis. Dev. Cell 6, 205–218 (2004)
Sakai, Y. et al. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 15, 213–225 (2001)
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnol. 20, 87–90 (2002)
Acknowledgements
We thank S. Kitajima and E. Ikeno for their assistance in generating the Mesp2–venus knock-in mouse; M. Ikumi and Y. Takahashi for technical assistance and maintaining the mice used in this study; A. Gossler and R. Johnson for providing the Dll1and lunatic fringe knockout mice; and H. Hamada for CYP26a-null embryos. We also thank H. Takeda for critical reading of this manuscript. We are grateful to H. Tanaka for preparing purified Mesp2 protein. This work was supported by Grants-in-Aid for Science Research on Priority Areas (B), the Organized Research Combination System and National BioResource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Movie 1
Changes in the expression domain of Mesp2-venus. (MOV 3804 kb)
Supplementary Figures 1-3
A knock-in strategy to generate Mesp2-venus (Fig. 1); the change of expression pattern of Mesp2-venus in vivo (Fig, 2); and double fluorescent in situ hybridization for L-fng and Mesp2 (Fig. 3). (JPG 89 kb)
Supplementary Figure 4
L-fng expression and Notch activity oscillate in the posterior PSM, but do not arrest correctly in the anterior PSM in the absence of Mesp2. (JPG 149 kb)
Supplementary Figure 5 and 6
A schematic representation of the sequential changes in the expression patterns (Fig. 5); expression pattern of Mesp2 in CYP26a-null embryos (Fig. 6). (JPG 120 kb)
Supplementary Data
This document contains legends for Supplementary Movie 1 and Supplementary Figs 1-6, and Supplementary Methods and additional references. (DOC 27 kb)
Rights and permissions
About this article
Cite this article
Morimoto, M., Takahashi, Y., Endo, M. et al. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature 435, 354–359 (2005). https://doi.org/10.1038/nature03591
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03591
This article is cited by
-
Periodic formation of epithelial somites from human pluripotent stem cells
Nature Communications (2022)
-
Understanding paraxial mesoderm development and sclerotome specification for skeletal repair
Experimental & Molecular Medicine (2020)
-
Coupling delay controls synchronized oscillation in the segmentation clock
Nature (2020)
-
MESP2 variants contribute to conotruncal heart defects by inhibiting cardiac neural crest cell proliferation
Journal of Molecular Medicine (2020)
-
Notch and Hippo signaling converge on Strawberry Notch 1 (Sbno1) to synergistically activate Cdx2 during specification of the trophectoderm
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.