Abstract
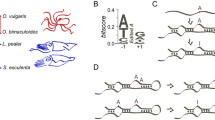
Most RNA editing systems are mechanistically diverse, informationally restorative, and scattershot in eukaryotic lineages1. In contrast, genetic recoding by adenosine-to-inosine RNA editing seems common in animals; usually, altering highly conserved or invariant coding positions in proteins2,3,4. Here I report striking variation between species in the recoding of synaptotagmin I (sytI). Fruitflies, mosquitoes and butterflies possess shared and species-specific sytI editing sites, all within a single exon. Honeybees, beetles and roaches do not edit sytI. The editing machinery is usually directed to modify particular adenosines by information stored in intron-mediated RNA structures5,6,7. Combining comparative genomics of 34 species with mutational analysis reveals that complex, multi-domain, pre-mRNA structures solely determine species-appropriate RNA editing. One of these is a previously unreported long-range pseudoknot. I show that small changes to intronic sequences, far removed from an editing site, can transfer the species specificity of editing between RNA substrates. Taken together, these data support a phylogeny of sytI gene editing spanning more than 250 million years of hexapod evolution. The results also provide models for the genesis of RNA editing sites through the stepwise addition of structural domains, or by short walks through sequence space from ancestral structures.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bass, B. L. (ed.) RNA Editing (Oxford University Press, Oxford, 2001)
Seeburg, P. & Hartner, J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 13, 279–283 (2003)
Hoopengardner, B., Bhalla, T., Staber, C. & Reenan, R. Nervous system targets of RNA editing identified by comparative genomics. Science 301, 832–836 (2003)
Bhalla, T., Rosenthal, J. J., Holmgren, M. & Reenan, R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nature Struct. Mol. Biol. 11, 950–956 (2004)
Herb, A., Higuchi, M., Sprengel, R. & Seeburg, P. Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc. Natl Acad. Sci. USA 93, 1875–1880 (1996)
Dawson, T., Sansam, C. & Emeson, R. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 279, 4941–4951 (2004)
Higuchi, M. et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75, 1361–1370 (1993)
Bass, B. L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 (2002)
Tonkin, L. A. et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans . EMBO J. 21, 6025–6035 (2002)
Palladino, M. J., Keegan, L. P., O'Connell, M. A. & Reenan, R. A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102, 437–449 (2000)
Higuchi, M. et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000)
Gurevich, I. et al. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34, 349–356 (2002)
Kawahara, Y. et al. Glutamate receptors: RNA editing and death of motor neurons. Nature 427, 801 (2004)
Brusa, R. et al. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270, 1677–1680 (1995)
Vissel, B. et al. The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron 29, 217–227 (2001)
Aronoff, R., Mellem, J. E., Maricq, A. V., Sprengel, R. & Seeburg, P. H. Neuronal toxicity in Caenorhabditis elegans from an editing site mutant in glutamate receptor channels. J. Neurosci. 24, 8135–8140 (2004)
Hanrahan, C. J., Palladino, M. J., Ganetzky, B. & Reenan, R. A. RNA editing of the Drosophila para Na+ channel transcript. Evolutionary conservation and developmental regulation. Genetics 155, 1149–1160 (2000)
Grauso, M., Reenan, R. A., Culetto, E. & Sattelle, D. B. Novel putative nicotinic acetylcholine receptor subunit genes, Dα5, Dα6 and Dα7, in Drosophila melanogaster identify a new and highly conserved target of adenosine deaminase acting on RNA-mediated A-to-I pre-mRNA editing. Genetics 160, 1519–1533 (2002)
Lehmann, K. A. & Bass, B. L. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 291, 1–13 (1999)
Yoshihara, M. & Littleton, J. T. Synaptotagmin I functions as a calcium sensor to synchronize neurotransmitter release. Neuron 36, 897–908 (2002)
Craxton, M. Genomic analysis of synaptotagmin genes. Genomics 77, 43–49 (2001)
Mackler, J. M. & Reist, N. E. Mutations in the second C2 domain of synaptotagmin disrupt synaptic transmission at Drosophila neuromuscular junctions. J. Comp. Neurol. 436, 4–16 (2001)
Chapman, E. R., Desai, R. C., Davis, A. F. & Tornehl, C. K. Delineation of the oligomerization, AP-2 binding, and synprint binding region of the C2B domain of synaptotagmin. J. Biol. Chem. 273, 32966–32972 (1998)
Rickman, C. et al. Synaptotagmin interaction with the syntaxin/SNAP-25 dimer is mediated by an evolutionarily conserved motif and is sensitive to inositol hexakisphosphate. J. Biol. Chem. 279, 12574–12579 (2004)
Grass, I., Thiel, S., Honing, S. & Haucke, V. Recognition of a basic AP-2 binding motif within the C2B domain of synaptotagmin is dependent on multimerization. J. Biol. Chem. 279, 54872–54880 (2004)
Nakhost, A., Houeland, G., Blandford, V. E., Castellucci, V. F. & Sossin, W. S. Identification and characterization of a novel C2B splice variant of synaptotagmin I. J. Neurochem. 89, 354–363 (2004)
Reenan, R., Hanrahan, C. & Ganetzky, B. The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron 25, 139–149 (2000)
Aruscavage, P. & Bass, B. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA 6, 257–269 (2000)
Kung, S. S., Chen, Y. C., Lin, W. H., Chen, C. C. & Chow, W. Y. Q/R RNA editing of the AMPA receptor subunit 2 (GRIA2) transcript evolves no later than the appearance of cartilaginous fishes. FEBS Lett. 509, 277–281 (2001)
Wheeler, W. C., Whiting, M., Wheeler, Q. D. & Carpernter, J. M. The phylogeny of the extant hexapod orders. Cladistics 17, 113–169 (2001)
Acknowledgements
I thank L. Reenan for discussions; B. Hoopengardner, T. Bhalla and A. Das for comments on the manuscript; B. Hoopengardner for sharing certain genomic DNA templates and for assistance with S2 cell culture; UCHC Molecular Core Facility staff for diligent sequencing efforts; and M. Lalande for his encouragement. This work was supported by grants from the National Science Foundation and National Institutes of Health (R.A.R.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares that he has no competing financial interests.
Supplementary information
Supplementary Methods
This document contains methods sections: 1. Insects used in this study. 2. Cloning and sequencing of synaptotagmin orthologues. 3. Primers used in this study. (PDF 117 kb)
Supplementary Legends
This document contains the legends for Supplementary Figures S1 through S5. (PDF 66 kb)
Supplementary Table S1
Contains all species information on insects and molecular data on sytI editing status in tabular form. (PDF 74 kb)
Supplementary Figure S1
Sequence alignment of synaptotagmin I from Drosophila species. (PDF 3162 kb)
Supplementary Figure S2
Sequence alignment of synaptotagmin I from Lepidopteran species. (PDF 2694 kb)
Supplementary Figure S3
Variation of synaptotagmin I editing structure within Lepidoptera. (PDF 1209 kb)
Supplementary Figure S4
Sequence alignment of synaptotagmin I from Anopheles species. (PDF 2756 kb)
Supplementary Figure S5
Comparison of Drosophila and Anopheles pseudoknot structure. (PDF 322 kb)
Rights and permissions
About this article
Cite this article
Reenan, R. Molecular determinants and guided evolution of species-specific RNA editing. Nature 434, 409–413 (2005). https://doi.org/10.1038/nature03364
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03364
This article is cited by
-
Dissecting the basis for differential substrate specificity of ADAR1 and ADAR2
Nature Communications (2023)
-
A hierarchy in clusters of cephalopod mRNA editing sites
Scientific Reports (2022)
-
Aberrant Overexpression of RNA-Editing Enzyme ADAR1 Promotes the Progression of Endometriosis
Reproductive Sciences (2020)
-
Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them
Nature Reviews Molecular Cell Biology (2018)
-
A-to-I RNA editing — immune protector and transcriptome diversifier
Nature Reviews Genetics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.