Abstract
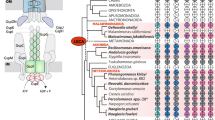
Hydrogenosomes are double-membraned ATP-producing and hydrogen-producing organelles of diverse anaerobic eukaryotes1. In some versions of endosymbiotic theory they are suggested to be homologues of mitochondria2,3,4, but alternative views suggest they arose from an anaerobic bacterium that was distinct from the mitochondrial endosymbiont5,6. Here we show that the 51-kDa and 24-kDa subunits of the NADH dehydrogenase module in complex I, the first step in the mitochondrial respiratory chain7, are active in hydrogenosomes of Trichomonas vaginalis. Like mitochondrial NADH dehydrogenase, the purified Trichomonas enzyme can reduce a variety of electron carriers including ubiquinone, but unlike the mitochondrial enzyme it can also reduce ferredoxin, the electron carrier used1 for hydrogen production. The presence of NADH dehydrogenase solves the long-standing conundrum of how hydrogenosomes regenerate NAD+ after malate oxidation. Phylogenetic analyses show that the Trichomonas 51-kDa homologue shares common ancestry with the mitochondrial enzyme. Recruitment of complex I subunits into a H2-producing pathway provides evidence that mitochondria and hydrogenosomes are aerobic and anaerobic homologues of the same endosymbiotically derived organelle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Müller, M. The hydrogenosome. J. Gen. Microbiol. 139, 2879–2889 (1993)
Embley, T. M., Horner, D. S. & Hirt, R. P. Anaerobic eukaryote evolution: hydrogenosomes as biochemically modified mitochondria? Trends Ecol. Evol. 12, 437–441 (1997)
Martin, W. & Müller, M. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41 (1998)
Embley, T. M. et al. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life 55, 387–395 (2003)
Whatley, J. M., John, P. & Whatley, F. R. From extracellular to intracellular: the establishment of mitochondria and chloroplasts. Proc. R. Soc. Lond. B 204, 165–187 (1979)
Dyall, S. D., Brown, M. T. & Johnson, P. J. Ancient invasions: from endosymbionts to organelles. Science 304, 253–257 (2004)
Yano, T. The energy-transducing NADH: quinone oxidoreductase, complex I. Mol. Aspects Med. 23, 345–368 (2002)
Clemens, D. L. & Johnson, P. J. Failure to detect DNA in hydrogenosomes of Trichomonas vaginalis by nick translation and immunomicroscopy. Mol. Biol. Parasitol. 106, 307–313 (2000)
Benchimol, M., Johnson, P. J. & deSouza, W. Morphogenesis of the hydrogenosome: An ultrastructural study. Biol. Cell 87, 197–205 (1996)
Sutak, R. et al. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl Acad. Sci. USA 101, 10368–10373 (2004)
Biagini, G. A., Finlay, B. J. & Lloyd, D. Evolution of the hydrogenosome. FEMS Microbiol. Lett. 155, 133–140 (1997)
Dyall, S. D. et al. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol. Cell. Biol. 20, 2488–2497 (2000)
Plumper, E., Bradley, P. J. & Johnson, P. J. Implications of protein import on the origin of hydrogenosomes. Protist 149, 303–311 (1998)
Drmota, T. et al. Iron-ascorbate cleavable malic enzyme from hydrogenosomes of Trichomonas vaginalis: purification and characterization. Mol. Biochem. Parasitol. 83, 221–234 (1996)
Whelan, S. & Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 (2001)
Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002)
Hendy, M. D. & Penny, D. A framework for the quantitative study of evolutionary trees. Syst. Zool. 38, 297–309 (1989)
Foster, P. G. Modeling compositional heterogeneity. Syst. Biol. 53, 485–495 (2004)
Dayhoff, M. O., Schwartz, R. M. & Orcutt, B. C. in Atlas of Protein Sequences and Structure (ed. Dayhoff, M. O.) 345–352 (National Biomedical Research Foundation, Washington DC, 1978)
Martin, W. et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165 (1998)
Delsuc, F., Phillips, M. J. & Penny, D. Comment on ‘Hexapod origins: monophyletic or paraphyletic?’. Science 301, 1482–1483 (2003)
Steinbuchel, A. & Muller, M. Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol. Biochem. Parasitol. 20, 57–65 (1986)
Rasoloson, D. et al. Mechanisms of in vitro development of resistance to metronidazole in Trichomonas vaginalis. Microbiology 148, 2467–2477 (2002)
Pilkington, S. J., Skehel, J. M., Gennis, R. B. & Walker, J. E. Relationship between mitochondrial NADH-ubiquinone reductase and a bacterial NAD-reducing hydrogenase. Biochemistry 30, 2166–2175 (1991)
Owen, R. Lectures on the Comparative Physiology of the Invertebrate Animals (Longman, Brown, Green, Longmans, London, 1843)
Embley, T. M. et al. Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc. R. Soc. Lond. B 262, 87–93 (1995)
Rotte, C., Stejskal, F., Zhu, G., Keithly, J. S. & Martin, W. Pyruvate: NADP oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: A biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol. Biol. Evol. 18, 710–720 (2001)
Tachezy, J., Sanchez, L. B. & Muller, M. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol. Biol. Evol. 18, 1919–1928 (2001)
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003)
Gaziova, I. & Lukes, J. Mitochondrial and nuclear localization of topoisomerase II in the flagellate Bodo saltans (Kinetoplastida), a species with non-catenated kinetoplast DNA. J. Biol. Chem. 278, 10900–10907 (2003)
Acknowledgements
We thank M. Hubalek for mass spectrometry and P. Dyal for technical support. Sequence data for Trichomonas vaginalis were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of T. vaginalis was accomplished with support from The National Institute of Allergy and Infectious Diseases. This work was supported by a Fogarty International Research Collaboration Award to J.T. and Miklos Muller and a grant from the Grant Agency of the Czech Republic to J.T. R.P.H. was supported by a Wellcome Trust University Award.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
Diagram of the 5 supplementary phylogenetic trees listed in the Table S1 found in the below Supplementary Materials. (PDF 265 kb)
Supplementary Material
Supplementary Tables (S1–S4), and Supplementary Methods for enzyme assays and purification. (DOC 41 kb)
Rights and permissions
About this article
Cite this article
Hrdy, I., Hirt, R., Dolezal, P. et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432, 618–622 (2004). https://doi.org/10.1038/nature03149
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03149
This article is cited by
-
The iron-sulfur scaffold protein HCF101 unveils the complexity of organellar evolution in SAR, Haptista and Cryptista
BMC Ecology and Evolution (2021)
-
Phylogenomics provides robust support for a two-domains tree of life
Nature Ecology & Evolution (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.