Abstract
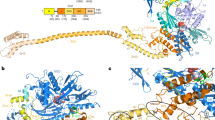
During protein biosynthesis, nascent polypeptide chains that emerge from the ribosomal exit tunnel encounter ribosome-associated chaperones, which assist their folding to the native state1,2. Here we present a 2.7 Å crystal structure of Escherichia coli trigger factor, the best-characterized chaperone of this type, together with the structure of its ribosome-binding domain in complex with the Haloarcula marismortui large ribosomal subunit. Trigger factor adopts a unique conformation resembling a crouching dragon with separated domains forming the amino-terminal ribosome-binding ‘tail’, the peptidyl-prolyl isomerase ‘head’, the carboxy-terminal ‘arms’ and connecting regions building up the ‘back’. From its attachment point on the ribosome, trigger factor projects the extended domains over the exit of the ribosomal tunnel, creating a protected folding space where nascent polypeptides may be shielded from proteases and aggregation. This study sheds new light on our understanding of co-translational protein folding, and suggests an unexpected mechanism of action for ribosome-associated chaperones.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bukau, B., Deuerling, E., Pfund, C. & Craig, E. A. Getting newly synthesized proteins into shape. Cell 101, 119–122 (2000)
Hartl, F. U. & Hayer-Hartl, M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 (2002)
Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002)
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000)
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000)
Hardesty, B. & Kramer, G. Folding of a nascent peptide on the ribosome. Prog. Nucleic Acid Res. Mol. Biol. 66, 41–66 (2001)
Agashe, V. R. et al. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117, 199–209 (2004)
Netzer, W. J. & Hartl, F. U. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349 (1997)
Nicola, A. V., Chen, W. & Helenius, A. Co-translational folding of an alphavirus capsid protein in the cytosol of living cells. Nature Cell Biol. 1, 341–345 (1999)
Kramer, G. et al. L23 protein functions as a chaperone docking site on the ribosome. Nature 419, 171–174 (2002)
Deuerling, E., Schulze-Specking, A., Tomoyasu, T., Mogk, A. & Bukau, B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696 (1999)
Teter, S. A. et al. Polypeptide flux through bacterial Hsp70: DnaK cooperates with Trigger Factor in chaperoning nascent chains. Cell 97, 755–765 (1999)
Hesterkamp, T., Deuerling, E. & Bukau, B. The amino-terminal 118 amino acids of Escherichia coli trigger factor constitute a domain that is necessary and sufficient for binding to ribosomes. J. Biol. Chem. 272, 21865–21871 (1997)
Kristensen, O. & Gajhede, M. Chaperone binding at the ribosomal exit tunnel. Structure (Camb.) 11, 1547–1556 (2003)
Vogtherr, M. et al. NMR solution structure and dynamics of the peptidyl-prolyl cis-trans isomerase domain of the trigger factor from Mycoplasma genitalium compared to FK506-binding protein. J. Mol. Biol. 318, 1097–1115 (2002)
Bitto, E. & McKay, D. B. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J. Biol. Chem. 278, 49316–49322 (2003)
Kramer, G. et al. Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains. J. Bacteriol. 186, 3777–3784 (2004)
Kramer, G. et al. Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli. J. Biol. Chem. 279, 14165–14170 (2004)
Behrens, S., Maier, R., de Cock, H., Schmid, F. X. & Gross, C. A. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20, 285–294 (2001)
Patzelt, H. et al. Three-state equilibrium of Escherichia coli trigger factor. Biol. Chem. 383, 1611–1619 (2002)
Hesterkamp, T., Hauser, S., Lütcke, H. & Bukau, B. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc. Natl Acad. Sci. USA 93, 4437–4441 (1996)
Maier, R., Eckert, B., Scholz, C., Lilie, H. & Schmid, F. X. Interaction of trigger factor with the ribosome. J. Mol. Biol. 326, 585–592 (2003)
Schatz, G. & Dobberstein, B. Common principles of protein translocation across membranes. Science 271, 1519–1526 (1996)
Beckmann, R. et al. Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science 278, 2123–2126 (1997)
Halic, M. et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814 (2004)
Pool, M. R., Stumm, J., Fulga, T. A., Sinning, I. & Dobberstein, B. Distinct modes of signal recognition particle interaction with the ribosome. Science 297, 1345–1348 (2002)
Otwinowski, Z. M. W. in Macromolecular Crystallography Part A Vol. 276 (ed. Carter, C. W. Jr, S.R.M.) 307–326 (Academic, San Diego, 1997)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999)
Jones, T. A., Bergdoll, M. & Kjeldgaard, O. in Crystallographic and Modeling Methods in Molecular Design (eds Bugg, C. & Ealick, S.) 189–195 (Springer-Verlag, New York, 1990)
Acknowledgements
Data collection was performed at the Swiss Light Source, Paul Scherrer Institut, Villigen and at the Swiss Norwegian Beamline (ESRF, Grenoble). We are grateful to C. Schulze-Briese, T. Tomizaki and A. Wagner at the SLS as well as P. Pattison and S. Capelli at the SNBL whose outstanding efforts have made these experiments possible. We also thank our colleagues S. Antolic, M. Steiner and members of the Ban laboratory for help in ribosome preparations and suggestions; and members of the Bukau laboratory for discussions. This work was supported by the Swiss National Science Foundation (SNSF), the NCCR Structural Biology program of the SNSF, an ETH research grant, grants of the Deutsche Forschungsgemeinschaft to B.B. and E.D. and a Young Investigator grant from the Human Frontier Science Program to N.B. and E.D.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
Example of experimental electron density quality. (JPG 122 kb)
Supplementary Figure 2
Structural flexibility of the Trigger Factor. (JPG 105 kb)
Supplementary Figure 3
Crystal contact sites of the full-length Trigger Factor. (JPG 59 kb)
Supplementary Figure 4
E.coli Trigger Factor associates with Haloarcula marismortui 50S and crosslinks to the ribosomal protein L23. (JPG 83 kb)
Supplementary Figure 5
Sequence conservation of L23 and ribosomal RNA at the TF binding site. (JPG 58 kb)
Supplementary Table
Data collection and refinement statistics. (DOC 42 kb)
Rights and permissions
About this article
Cite this article
Ferbitz, L., Maier, T., Patzelt, H. et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596 (2004). https://doi.org/10.1038/nature02899
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02899
This article is cited by
-
Cyclophilin anaCyp40 regulates photosystem assembly and phycobilisome association in a cyanobacterium
Nature Communications (2022)
-
Self-assembled raccoon dog parvovirus VP2 protein confers immunity against RDPV disease in raccoon dogs: in vitro and in vivo studies
Virology Journal (2021)
-
Functional cooperativity between the trigger factor chaperone and the ClpXP proteolytic complex
Nature Communications (2021)
-
A Review: Molecular Chaperone-mediated Folding, Unfolding and Disaggregation of Expressed Recombinant Proteins
Cell Biochemistry and Biophysics (2021)
-
Molecular chaperones and their denaturing effect on client proteins
Journal of Biomolecular NMR (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.