Abstract
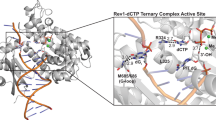
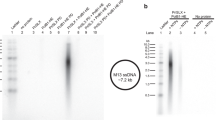
The 3′-terminal CCA nucleotide sequence (positions 74–76) of transfer RNA is essential for amino acid attachment1 and interaction with the ribosome2,3,4 during protein synthesis. The CCA sequence is synthesized de novo and/or repaired by a template-independent RNA polymerase, ‘CCA-adding enzyme’, using CTP and ATP as substrates5. Despite structural and biochemical studies5,6,7,8, the mechanism by which the CCA-adding enzyme synthesizes the defined sequence without a nucleic acid template remains elusive. Here we present the crystal structure of Aquifex aeolicus CCA-adding enzyme, bound to a primer tRNA lacking the terminal adenosine and an incoming ATP analogue, at 2.8 Å resolution. The enzyme enfolds the acceptor T helix of the tRNA molecule. In the catalytic pocket, C75 is adjacent to ATP, and their base moieties are stacked. The complementary pocket for recognizing C74-C75 of tRNA forms a ‘protein template’ for the penultimate two nucleotides, mimicking the nucleotide template used by template-dependent polymerases. These results are supported by systematic analyses of mutants. Our structure represents the ‘pre-insertion’ stage of selecting the incoming nucleotide and provides the structural basis for the mechanism underlying template-independent RNA polymerization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sprinzl, M. & Cramer, F. The -C-C-A end of tRNA and its role in protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 22, 1–69 (1979)
Green, R. & Noller, H. F. Ribosomes and translation. Annu. Rev. Biochem. 66, 679–716 (1997)
Kim, D. F. & Green, R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4, 859–864 (1999)
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000)
Deutscher, M. P. in Enzymes of Nucleic Acid Synthesis and Modification. RNA Enzymes vol. 2 (ed. Jacob, S. T.) 159–183 (CRC, Boca Raton, Fl, 1983)
Yue, D., Weiner, A. M. & Maizels, N. The CCA-adding enzyme has a single active site. J. Biol. Chem. 273, 29693–29700 (1998)
Shi, P. Y., Maizels, N. & Weiner, A. M. CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J. 17, 3197–3206 (1998)
Hou, Y. M. Unusual synthesis by the Escherichia coli CCA-adding enzyme. RNA 6, 1031–1043 (2000)
Holm, L. & Sander, C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 20, 345–347 (1995)
Martin, G. & Keller, W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 15, 2593–2603 (1996)
Yue, D., Maizels, N. & Weiner, A. M. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 2, 895–908 (1996)
Li, F. et al. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell 111, 815–824 (2002)
Okabe, M. et al. Divergent evolutions of trinucleotide polymerization revealed by an archaeal CCA-adding enzyme structure. EMBO J. 22, 5918–5927 (2003)
Xiong, Y., Li, F., Wang, J., Weiner, A. M. & Steitz, T. A. Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes. Mol. Cell 12, 1165–1172 (2003)
Tomita, K. & Weiner, A. M. Collaboration between CC- and A-adding enzymes to build and repair the 3′-terminal CCA of tRNA in Aquifex aeolicus. Science 294, 1334–1336 (2001)
Tomita, K. & Weiner, A. M. Closely related CC- and A-adding enzymes collaborate to construct and repair the 3′-terminal CCA of tRNA in Synechocystis sp. and Deinococcus radiodurans. J. Biol. Chem. 277, 48192–48198 (2003)
Augustin, M. A. et al. Crystal structure of the human CCA-adding enzyme: insights into template-independent polymerization. J. Mol. Biol. 328, 985–994 (2003)
Brautigam, C. A. & Steitz, T. A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8, 54–63 (1998)
Hegg, L. A. & Thurlow, D. L. Cytidines in tRNAs that are required intact by ATP/CTP:tRNA nucleotidyltransferases from Escherichia coli and Saccharomyces cerevisiae. Nucleic Acids Res. 18, 5975–9597 (1990)
Xiong, Y. & Steitz, T. A. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature doi:10.1038/nature02711 (this issue)
Shi, P. Y., Weiner, A. M. & Maizels, N. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA 4, 276–284 (1998)
Temiakov, D. et al. Structural basis for substrate selection by T7 RNA polymerase. Cell 116, 381–391 (2004)
Yin, Y. W. & Steitz, T. A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116, 393–404 (2004)
Pelletier, H., Sawaya, M. R., Kumar, A., Wilson, S. H. & Kraut, J. Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science 264, 1891–1903 (1994)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Weeks, C. M. & Miller, R. The design and implementation of SnB version 2.0. J. Appl. Crystallogr. 32, 120–124 (1999)
Collaborative Computational Project Number 4, The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Rice, L. M. & Brunger, A. T. Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins Struct. Funct. Genet. 19, 277–290 (1994)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Acknowledgements
We thank A. M. Weiner for the Aa.LC plasmids; and M. Kawamoto and H. Sakai for help with data collection at SPring-8. This work was supported by Kurata Memorial Hitachi Science and Technology Foundation, Takeda Science Foundation, Foundation of Advanced Technology Institute and a Grant-in-aid for Young Scientists (to K.T.); by Asahi Glass Foundation (to S.F.); by a grant from the Ministry of Education, Culture, Sports, Science and Technology (to N.T.); and by a PRESTO Program grant from Japan Science and Technology and a Naito Foundation grant (to O.N.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Table 1
Data collection and refinement statistics (DOC 56 kb)
Rights and permissions
About this article
Cite this article
Tomita, K., Fukai, S., Ishitani, R. et al. Structural basis for template-independent RNA polymerization. Nature 430, 700–704 (2004). https://doi.org/10.1038/nature02712
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02712
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.