Abstract
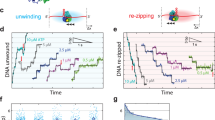
The NS3 helicase is essential for cytoplasmic RNA replication by the hepatitis C virus1,2,3,4, and it is a representative member of helicase superfamily 2 (SF2). NS3 is an important model system for understanding unwinding activities of DExH/D proteins5,6,7, and it has been the subject of extensive structural and mutational analyses8,9,10,11. Despite intense interest in NS3, the molecular and kinetic mechanisms for RNA unwinding by this helicase have remained obscure. We have developed a combinatorial, time-resolved approach for monitoring the microscopic behaviour of a helicase at each nucleotide of a duplex substrate. By applying this analysis to NS3, we have independently established the ‘physical’ and ‘kinetic’ step size for unwinding of RNA (18 base pairs, in each case), which we relate to the stoichiometry of the functional, translocating species. Having obtained microscopic unwinding rate constants at each position along the duplex, we demonstrate that NS3 unwinds RNA through a highly coordinated cycle of fast ripping and local pausing that occurs with regular spacing along the duplex substrate, much like the stepping behaviour of cytoskeletal motor proteins12.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Moradpour, D. et al. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 60, 103–109 (2003)
Dimitrova, M., Imbert, I., Kieny, M. P. & Schuster, C. Protein–protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77, 5401–5414 (2003)
Piccininni, S. et al. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 277, 45670–45679 (2002)
Gallinari, P. et al. Modulation of hepatitis C virus NS3 protease and helicase activities through the interaction with NS4A. Biochemistry 38, 5620–5632 (1999)
Bird, L. E., Subramanya, H. S. & Wigley, D. B. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8, 14–18 (1998)
Staley, J. P. & Guthrie, C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326 (1998)
Caruthers, J. M. & McKay, D. B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 (2002)
Kim, J. L. et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6, 89–100 (1998)
Kim, D. W., Kim, J., Gwack, Y., Han, J. H. & Choe, J. Mutational analysis of the hepatitis C virus RNA helicase. J. Virol. 71, 9400–9409 (1997)
Cho, H.-S. et al. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 273, 15045–15052 (1998)
Korolev, S., Yao, N., Lohman, T. M., Weber, P. C. & Waksman, G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 7, 605–610 (1998)
Vale, R. D. & Milligan, R. A. The way things move: looking under the hood of molecular motor proteins. Science 288, 88–95 (2000)
Bartenschlager, R. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepat. 6, 165–181 (1999)
Gallinari, P. et al. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J. Virol. 72, 6758–6769 (1998)
Pang, P. S., Jankowsky, E., Planet, P. J. & Pyle, A. M. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21, 1168–1176 (2002)
Paolini, C., De Francesco, R. & Gallinari, P. Enzymatic properties of hepatitis C virus NS3-associated helicase. J. Gen. Virol. 81, 1335–1345 (2000)
Lucius, A. L., Maluf, N. K., Fischer, C. J. & Lohman, T. M. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85, 2224–2239 (2003)
Ali, J. A. & Lohman, T. M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275, 377–380 (1997)
Bianco, P. R. & Kowalczykowski, S. C. Translocation step size and mechanism of the RecBC DNA helicase. Nature 405, 368–372 (2000)
Jankowsky, E., Gross, C. H., Shuman, S. & Pyle, A. M. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature 403, 447–451 (2000)
Bianco, P. R. et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 (2001)
Ha, T. et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419, 638–641 (2002)
Spies, M. et al. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell 114, 647–654 (2003)
Dessinges, M. N., Lionnet, T., Xi, X. G., Bensimon, D. & Croquette, V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl Acad. Sci. USA 101, 6439–6444 (2004)
Lucius, A. L. et al. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J. Mol. Biol. 324, 409–428 (2002)
Levin, M. K. & Patel, S. S. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274, 31839–31846 (1999)
Locatelli, G. A., Spadari, S. & Maga, G. Hepatitis C virus NS3 ATPase/helicase: an ATP switch regulates the cooperativity among the different substrate binding sites. Biochemistry 41, 10332–10342 (2002)
Levin, M. K., Wang, Y. H. & Patel, S. S. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279, 26005–26012 (2004)
Kawaoka, J., Jankowsky, E. & Pyle, A. M. Backbone tracking by the SF2 helicase NPH-II. Nature Struct. Mol. Biol. 11, 526–530 (2004)
Kuzmic, P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237, 260–273 (1996)
Acknowledgements
The authors would like to thank E. Jankowsky, J. Kawaoka and M. Brenowitz for discussions. We thank R. Beran for sharing his results on NS3 unwinding of multi-piece RNA substrates, and H. Le for discussions and early gifts of NS3 protein. This work was supported by grants from the NIH and the Howard Hughes Medical Institute. A.M.P. is an HHMI investigator.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
This figure describes NS3 titration experiments designed to determine functional oligomeric state of NS3: total NS3 binding to an RNA substrate as determined by filter binding assay is compared to functional binding (as monitored by the resulting unwinding amplitude). (PDF 13 kb)
Supplementary Figure 2
This figure shows model discrimination analysis of RNS unwinding. Timecourses for each RNS duplex length were fit to a simple sequential kinetic model (Scheme 1, Supplementary Methods), where the number of steps varied from 1 to 8. As shown on the figure, the squared residuals for each fit were plotted versus number of steps. The absolute minimum of squared residuals determines the number of rate-limiting steps for an RNS duplex of given length. (PDF 112 kb)
Supplementary Figure 3
Data from Supplementary Figure 2 are represented using color coding. Red color corresponds to the best fit for the total number of rate-limiting steps for each RNS duplex length. (PDF 8 kb)
Supplementary Figure 4
This figure shows four kinetic timecourses for NS3 unwinding of RNS duplexes within the same 18 bp kinetic step. The kinetic curves are virtually superimposable, although a small progressive deviation is observed for increasingly longer duplexes. (PDF 52 kb)
Supplementary Figure 5
This figure shows determination of physical step size and local processivity for an RNS duplex with a longer 3’-overhang (33 nt). The longer overhang does not change the pattern of local processivity along the duplex length. (PDF 86 kb)
Supplementary Methods
Supplementary Methods section includes extended description of kinetic analysis and global fitting for RNS unwinding by NS3. (DOC 35 kb)
Rights and permissions
About this article
Cite this article
Serebrov, V., Pyle, A. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430, 476–480 (2004). https://doi.org/10.1038/nature02704
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02704
This article is cited by
-
Fork sensing and strand switching control antagonistic activities of RecQ helicases
Nature Communications (2013)
-
An overview of HCV molecular biology, replication and immune responses
Virology Journal (2011)
-
BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation
The EMBO Journal (2009)
-
Non-hexameric DNA helicases and translocases: mechanisms and regulation
Nature Reviews Molecular Cell Biology (2008)
-
Structural basis for DNA duplex separation by a superfamily-2 helicase
Nature Structural & Molecular Biology (2007)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.