Abstract
Smooth muscle cells switch between differentiated and proliferative phenotypes in response to extracellular cues1, but the transcriptional mechanisms that confer such phenotypic plasticity remain unclear. Serum response factor (SRF) activates genes involved in smooth muscle differentiation and proliferation by recruiting muscle-restricted cofactors, such as the transcriptional coactivator myocardin, and ternary complex factors (TCFs) of the ETS-domain family, respectively2,3,4,5,6,7,8,9. Here we show that growth signals repress smooth muscle genes by triggering the displacement of myocardin from SRF by Elk-1, a TCF that acts as a myogenic repressor. The opposing influences of myocardin and Elk-1 on smooth muscle gene expression are mediated by structurally related SRF-binding motifs that compete for a common docking site on SRF. A mutant smooth muscle promoter, retaining responsiveness to myocardin and SRF but defective in TCF binding, directs ectopic transcription in the embryonic heart, demonstrating a role for TCFs in suppression of smooth muscle gene expression in vivo. We conclude that growth and developmental signals modulate smooth muscle gene expression by regulating the association of SRF with antagonistic cofactors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Owens, G. K. Regulation of differentiation of vascular smooth muscle cells. Physiol. Rev. 75, 487–517 (1995)
Chang, D. F. et al. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4, 107–118 (2003)
Wang, D. et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105, 851–862 (2001)
Du, K. L. et al. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell. Biol. 23, 2425–2437 (2003)
Chen, J., Kitchen, C. M., Streb, J. W. & Miano, J. M. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell. Cardiol. 34, 1345–1356 (2002)
Yoshida, T. et al. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ. Res. 92, 856–864 (2003)
Wang, Z., Wang, D. Z., Pipes, G. C. & Olson, E. N. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl Acad. Sci. USA 100, 7129–7134 (2003)
Li, S., Wang, D. Z., Wang, Z., Richardson, J. A. & Olson, E. N. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl Acad. Sci. USA 100, 9366–9370 (2003)
Treisman, R. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4, 96–101 (1994)
Miano, J. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 35, 577–593 (2003)
Shaw, P. E., Schroter, H. & Nordheim, A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell 56, 563–572 (1989)
Giovane, A., Pintzas, A., Maira, S. M., Sobieszczuk, P. & Wasylyk, B. Net, a new ets transcription factor that is activated by Ras. Genes Dev. 8, 1502–1513 (1994)
Sharrocks, A. D. Complexities in ETS-domain transcription factor function and regulation: lessons from the TCF (ternary complex factor) subfamily. The Colworth Medal Lecture. Biochem. Soc. Trans. 30, 1–9 (2002)
Janknecht, R., Ernst, W. H., Pingoud, V. & Nordheim, A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 12, 5097–5104 (1993)
Marais, R., Wynne, J. & Treisman, R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73, 381–393 (1993)
Holycross, B. J., Blank, R. S., Thompson, M. M., Peach, M. J. & Owens, G. K. Platelet-derived growth factor-BB-induced suppression of smooth muscle cell differentiation. Circ. Res. 71, 1525–1532 (1992)
Shore, P. et al. Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol. Cell. Biol. 16, 3338–3349 (1996)
Li, L., Liu, Z., Mercer, B., Overbeek, P. & Olson, E. N. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev. Biol. 187, 311–321 (1997)
Kim, S., Ip, H. S., Lu, M. M., Clendenin, C. & Parmacek, M. S. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 17, 2266–2278 (1997)
Mack, C. P. & Owens, G. K. Regulation of smooth muscle alpha-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ. Res. 84, 852–861 (1999)
Heldin, C. H., Ostman, A. & Ronnstrand, L. Signal transduction via platelet-derived growth factor receptors. Biochim. Biophys. Acta 1378, F79–113 (1998)
Ling, Y., West, A. G., Roberts, E. C., Lakey, J. H. & Sharrocks, A. D. Interaction of transcription factors with serum response factor. Identification of the Elk-1 binding surface. J. Biol. Chem. 273, 10506–10514 (1998)
Miralles, F., Posern, G., Zaromytidou, A. I. & Treisman, R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113, 329–342 (2003)
Ling, Y., Lakey, J. H., Roberts, C. E. & Sharrocks, A. D. Molecular characterization of the B-box protein-protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J. 16, 2431–2440 (1997)
Hassler, M. & Richmond, T. J. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 20, 3018–3028 (2001)
Li, L., Miano, J. M., Cserjesi, P. & Olson, E. N. SM22, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ. Res. 78, 188–195 (1996)
Cen, B. et al. Megakaryoblastic leukemia 1, a potent transcriptional activator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 23, 6597–6608 (2003)
Wang, D. Z. et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl Acad. Sci. USA 99, 14855–14860 (2002)
Qiu, P. & Li, L. Histone acetylation and recruitment of serum responsive factor and CREB-binding protein onto SM22 promoter during SM22 gene expression. Circ. Res. 90, 858–865 (2002)
Acknowledgements
We thank A. Sharrocks, M. Parmacek and L. Schuger for reagents, A. Tizenor for graphics, J. Page for editorial assistance, L. Sutherland for technical assistance and R. Bassel-Duby, L. Li, M. Tallquist and H. Yanagisawa for discussions. This work was supported by grants from the NIH, the McGowan Foundation, the Donald W. Reynolds Foundation, and the Robert A. Welch Foundation to E.N.O., from the Muscular Dystrophy Association (MDA) and NIH to D.-Z.W., and by the DFG to A.N.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
Modulation of smooth muscle gene expression by PDGF. (JPG 42 kb)
Supplementary Figure 2
ChIP assay to detect association of myocardin and Elk-1 with the a-smooth muscle actin gene promoter. (JPG 35 kb)
Supplementary Figure 3
Gel mobility shift assays with the CArG and TCF binding sites from the c-fos and SM22 promoter. (JPG 64 kb)
Supplementary Figure 4
Loss of myogenic activity of myocardin mutant L291P. (JPG 26 kb)
Supplementary Figure 5
Phosphorylation of Elk-1 in the presence of serum and inhibition by U0126. (JPG 28 kb)
Supplementary Figure 6
Repression of myocardin transcriptional activity by Elk-1. (JPG 79 kb)
Rights and permissions
About this article
Cite this article
Wang, Z., Wang, DZ., Hockemeyer, D. et al. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428, 185–189 (2004). https://doi.org/10.1038/nature02382
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02382
This article is cited by
-
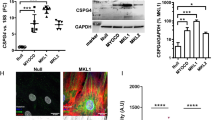
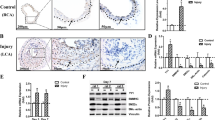
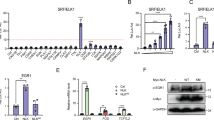
Vascular injury activates the ELK1/SND1/SRF pathway to promote vascular smooth muscle cell proliferative phenotype and neointimal hyperplasia
Cellular and Molecular Life Sciences (2024)
-
NCOR1 maintains the homeostasis of vascular smooth muscle cells and protects against aortic aneurysm
Cell Death & Differentiation (2023)
-
Progerin induces a phenotypic switch in vascular smooth muscle cells and triggers replication stress and an aging-associated secretory signature
GeroScience (2023)
-
SRF: a seriously responsible factor in cardiac development and disease
Journal of Biomedical Science (2022)
-
NLK is required for Ras/ERK/SRF/ELK signaling to tune skeletal muscle development by phosphorylating SRF and antagonizing the SRF/MKL pathway
Cell Death Discovery (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.