Abstract
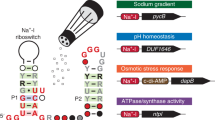
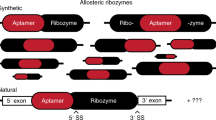
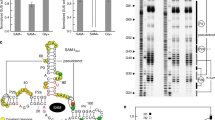
Most biological catalysts are made of protein; however, eight classes of natural ribozymes have been discovered that catalyse fundamental biochemical reactions. The central functions of ribozymes in modern organisms support the hypothesis that life passed through an ‘RNA world’ before the emergence of proteins and DNA. We have identified a new class of ribozymes that cleaves the messenger RNA of the glmS gene in Gram-positive bacteria. The ribozyme is activated by glucosamine-6-phosphate (GlcN6P), which is the metabolic product of the GlmS enzyme. Additional data indicate that the ribozyme serves as a metabolite-responsive genetic switch that represses the glmS gene in response to rising GlcN6P concentrations. These findings demonstrate that ribozyme switches may have functioned as metabolite sensors in primitive organisms, and further suggest that modern cells retain some of these ancient genetic control systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cech, T. R. Self-splicing of group I introns. Annu. Rev. Biochem. 59, 543–568 (1990)
Frank, D. N. & Pace, N. R. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67, 153–180 (1998)
Michel, F. & Feral, J. Structure and activities of group II introns. Annu. Rev. Biochem. 64, 435–461 (1995)
Doherty, E. A. & Doudna, J. A. Ribozyme structures and mechanisms. Annu. Rev. Biochem. 69, 597–615 (2000)
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000)
Breaker, R. R. In vitro selection of catalytic polynucleotides. Chem. Rev. 97, 371–390 (1997)
Osborne, S. E. & Ellington, A. D. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 97, 349–370 (1997)
Soukup, G. A. & Breaker, R. R. Allosteric nucleic acid catalysts. Curr. Opin. Struct. Biol. 10, 318–325 (2000)
Breaker, R. R. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 13, 31–39 (2002)
Silverman, S. K. Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA. RNA 9, 377–383 (2003)
Seetharaman, S., Zivarts, M., Sudarsan, N. & Breaker, R. R. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nature Biotechnol. 19, 336–341 (2001)
Ptashne, M. & Gann, A. Genes & Signals (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2002)
Nahvi, A. et al. Genetic control by a metabolite-binding mRNA. Chem. Biol. 9, 1043–1049 (2002)
Winkler, W., Nahvi, A. & Breaker, R. R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002)
Winkler, W. C., Cohen-Chalamish, S. & Breaker, R. R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA 99, 15908–15913 (2002)
Mironov, A. S. et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756 (2002)
Winkler, W. C. & Breaker, R. R. Genetic control by metabolite-binding riboswitches. Chembiochem 4, 1024–1032 (2003)
Mandal, M., Boese, B., Barrick, J. E., Winkler, W. C. & Breaker, R. R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 (2003)
Gusarov, I. & Nudler, E. The mechanism of intrinsic transcription termination. Mol. Cell 3, 495–504 (1999)
Yarnell, W. S. & Roberts, J. W. Mechanism of intrinsic transcription termination and antitermination. Science 284, 611–615 (1999)
Mandal, M. & Breaker, R. R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Struct. Mol. Biol. 11, 29–35 (2004)
Vitreschak, A. G., Rodionov, D. A., Mironov, A. A. & Gelfand, M. S. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 20, 44–50 (2004)
Sudarsan, N., Barrick, J. E. & Breaker, R. R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9, 644–647 (2003)
Kubodera, T. et al. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 555, 516–520 (2003)
Barrick, J. E. et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA (submitted)
Milewski, S. Glucosamine-6-phosphate synthase: the multi-facets enzyme. Biochim. Biophys. Acta 1597, 173–192 (2002)
Winkler, W. C., Nahvi, A., Sudarsan, N., Barrick, J. E. & Breaker, R. R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Struct. Biol. 10, 701–707 (2003)
Sudarsan, N., Wickiser, J. K., Nakamura, S., Ebert, M. S. & Breaker, R. R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 17, 2688–2697 (2003)
Soukup, G. A. & Breaker, R. R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999)
Li, Y. & Breaker, R. R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 121, 5364–5372 (1999)
Emilsson, G. M., Nakamura, S., Roth, A. & Breaker, R. R. Ribozyme speed limits. RNA 9, 907–918 (2003)
Breaker, R. R. et al. A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. RNA 9, 949–957 (2003)
Tang, J. & Breaker, R. R. Rational design of allosteric ribozymes. Chem. Biol. 4, 453–459 (1997)
Soukup, G. A. & Breaker, R. R. Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA 96, 3584–3589 (1999)
Koizumi, M., Soukup, G. A., Kerr, J. Q. & Breaker, R. R. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nature Struct. Biol. 6, 1062–1071 (1999)
Breaker, R. R. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 13, 31–39 (2002)
Soukup, G. A., DeRose, E. C., Koizumi, M. & Breaker, R. R. Generating new ligand-binding RNAs by affinity maturation and disintegration of allosteric ribozymes. RNA 7, 524–536 (2001)
Abrash, H. I., Cheung, C.-C. S. & Davis, J. C. The nonenzymic hydrolysis of nucleoside 2′,3′-phosphates. Biochemistry 6, 1303 (1967)
Saville, B. J. & Collins, R. A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 61, 685–696 (1990)
Joyce, G. F. The antiquity of RNA-based evolution. Nature 418, 214–221 (2002)
Miller, J. H. A Short Course in Bacterial Genetics 72 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1992)
Acknowledgements
We thank K. Corbino for generating the glmS IGR sequence alignments, and members of the Breaker laboratory for discussions. This work was supported by grants from the NIH and the NSF. R.R.B is also grateful for support from the Yale Liver Center and the David and Lucile Packard Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Winkler, W., Nahvi, A., Roth, A. et al. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004). https://doi.org/10.1038/nature02362
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02362
This article is cited by
-
CRISPR–dCas12a-mediated genetic circuit cascades for multiplexed pathway optimization
Nature Chemical Biology (2023)
-
Structure-based insights into recognition and regulation of SAM-sensing riboswitches
Science China Life Sciences (2023)
-
Structure and mechanism of the methyltransferase ribozyme MTR1
Nature Chemical Biology (2022)
-
Getting to the bottom of lncRNA mechanism: structure–function relationships
Mammalian Genome (2022)
-
Theoretical Studies of the Self Cleavage Pistol Ribozyme Mechanism
Topics in Catalysis (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.