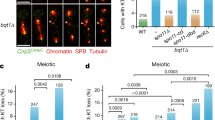
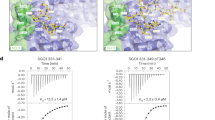
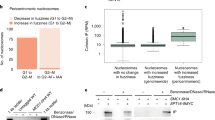
Abstract
Meiosis comprises a pair of specialized nuclear divisions that produce haploid germ cells. To accomplish this, sister chromatids must segregate together during the first meiotic division (meiosis I), which requires that sister chromatid cohesion persists at centromeres. The factors that protect centromeric cohesion during meiosis I have remained elusive. Here we identify Sgo1 (shugoshin), a protector of the centromeric cohesin Rec8 in fission yeast. We also identify a homologue of Sgo1 in budding yeast. We provide evidence that shugoshin is widely conserved among eukaryotes. Moreover, we identify Sgo2, a paralogue of shugoshin in fission yeast, which is required for faithful mitotic chromosome segregation. Localization of Sgo1 and Sgo2 at centromeres requires the kinase Bub1, identifying shugoshin as a crucial target for the kinetochore function of Bub1. These findings provide insights into the evolution of meiosis and kinetochore regulation during mitosis and meiosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nasmyth, K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673–745 (2001)
Koshland, D. E. & Guacci, V. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 12, 297–301 (2000)
Uhlmann, F. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 13, R104–R114 (2003)
Lee, J. Y. & Orr-Weaver, T. L. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17, 753–777 (2001)
Hirano, T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16, 399–414 (2002)
Klein, F. et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 (1999)
Parisi, S. et al. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family, conserved from fission yeast to humans. Mol. Cell. Biol. 19, 3515–3528 (1999)
Watanabe, Y. & Nurse, P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400, 461–464 (1999)
Pasierbek, P. et al. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15, 1349–1360 (2001)
Eijpe, M., Offenberg, H., Jessberger, R., Revenkova, E. & Heyting, C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J. Cell Biol. 160, 657–670 (2003)
Stoop-Myer, C. & Amon, A. Meiosis: Rec8 is the reason for cohesion. Nature Cell Biol. 1, E125–E127 (1999)
Buonomo, S. B. et al. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103, 387–398 (2000)
Kitajima, T. S., Miyazaki, Y., Yamamoto, M. & Watanabe, Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 22, 5643–5653 (2003)
Shonn, M. A., McCarroll, R. & Murray, A. W. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16, 1659–1671 (2002)
Lee, B. H., Amon, A. & Prinz, S. Spo13 regulates cohesin cleavage. Genes Dev. 16, 1672–1681 (2002)
Blower, M. D. & Karpen, G. H. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nature Cell Biol. 3, 730–739 (2001)
Kerrebrock, A. W., Moore, D. P., Wu, J. S. & Orr-Weaver, T. L. MEI-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83, 247–256 (1995)
Kitajima, T. S., Yokobayashi, S., Yamamoto, M. & Watanabe, Y. Distinct cohesin complexes organize meiotic chromosome domains. Science 300, 1152–1155 (2003)
Toth, A. et al. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103, 1155–1168 (2000)
Yokobayashi, S., Yamamoto, M. & Watanabe, Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 23, 3965–3973 (2003)
Yamamoto, A. & Hiraoka, Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 22, 2284–2296 (2003)
Mata, J., Lyne, R., Burns, G. & Bähler, J. The transcriptional program of meiosis and sporulation in fission yeast. Nature Genet. 32, 143–147 (2002)
Watanabe, Y., Yokobayashi, S., Yamamoto, M. & Nurse, P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409, 359–363 (2001)
Saitoh, S., Takahashi, K. & Yanagida, M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90, 131–143 (1997)
Ekwall, K. et al. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269, 1429–1431 (1995)
Bernard, P., Maure, J. F. & Javerzat, J. P. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nature Cell Biol. 3, 522–526 (2001)
Yamaguchi, S., Decottignies, A. & Nurse, P. Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint. EMBO J. 22, 1075–1087 (2003)
Bernard, P., Hardwick, K. & Javerzat, J. P. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143, 1775–1787 (1998)
Henikoff, S., Pietrokovski, S. & Henikoff, J. G. Superior performance in protein homology detection with the Blocks Database servers. Nucleic Acids Res. 26, 309–312 (1998)
Bailey, T. L. & Gribskov, M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14, 48–54 (1998)
Tang, T. T., Bickel, S. E., Young, L. M. & Orr-Weaver, T. L. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S322 protein. Genes Dev. 12, 3843–3856 (1998)
Kumada, K. et al. Cut1 is loaded onto the spindle by binding to Cut2 and promotes anaphase spindle movement upon Cut2 proteolysis. Curr. Biol. 8, 633–641 (1998)
LeBlanc, H. N., Tang, T. T., Wu, J. S. & Orr-Weaver, T. L. The mitotic centromeric protein MEI-S332 and its role in sister-chromatid cohesion. Chromosoma 108, 401–411 (1999)
Bernard, P. et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 (2001)
Nonaka, N. et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature Cell Biol. 4, 89–93 (2002)
Cahill, D. P. et al. Mutations of mitotic checkpoint genes in human cancers. Nature 392, 300–303 (1998)
Scanlan, M. J. et al. Humoral immunity to human breast cancer: antigen definition and quantitative analysis of mRNA expression. Cancer Immun. 1, 4 (2001)
Hassold, T. & Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nature Rev. Genet. 2, 280–291 (2001)
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998)
Rao, H., Uhlmann, F., Nasmyth, K. & Varshavsky, A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410, 955–959 (2001)
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998)
Rabitsch, K. P. et al. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell 4, 535–548 (2003)
Glynn, J. M., Lustig, R. J., Berlin, A. & Chang, F. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11, 836–845 (2001)
Lupas, A., Van Dyke, M. & Stock, J. Predicting coiled coils from protein sequences. Science 252, 1162–1164 (1991)
Acknowledgements
We thank J. P. Cooper for critical reading of the manuscript; S. Hauf, M. Ohsugi and R. Watanabe for suggestions; J. P. Javerzat, F. Chang, T. Toda, M. Yanagida and P. Nurse for strains and plasmids of fission yeast; and F. Klein, K. P. Rabitsch, K. Nasmyth, M. Longtine, A. Shinohara and T. Maeda for strains and methods of budding yeast. We appreciate the support of M. Yamamoto and all members of his laboratory for their help. This work was supported in part by grants from the Ministry of Education, Science and Culture of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Kitajima, T., Kawashima, S. & Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427, 510–517 (2004). https://doi.org/10.1038/nature02312
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02312
This article is cited by
-
ATM signaling modulates cohesin behavior in meiotic prophase and proliferating cells
Nature Structural & Molecular Biology (2023)
-
The Caenorhabditis elegans Shugoshin regulates TAC-1 in cilia
Scientific Reports (2023)
-
SGOL2 is a novel prognostic marker and fosters disease progression via a MAD2-mediated pathway in hepatocellular carcinoma
Biomarker Research (2022)
-
Centromeres are dismantled by foundational meiotic proteins Spo11 and Rec8
Nature (2021)
-
Methotrexate impaired in-vivo matured mouse oocyte quality and the possible mechanisms
BMC Molecular and Cell Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.