Abstract
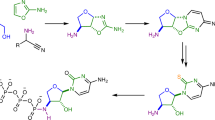
Fluorine is the thirteenth most abundant element in the earth's crust, but fluoride concentrations in surface water are low and fluorinated metabolites are extremely rare1,2. The fluoride ion is a potent nucleophile in its desolvated state, but is tightly hydrated in water and effectively inert. Low availability and a lack of chemical reactivity have largely excluded fluoride from biochemistry: in particular, fluorine's high redox potential precludes the haloperoxidase-type mechanism3,4 used in the metabolic incorporation of chloride and bromide ions. But fluorinated chemicals are growing in industrial importance, with applications in pharmaceuticals, agrochemicals and materials products5,6,7. Reactive fluorination reagents requiring specialist process technologies are needed in industry and, although biological catalysts for these processes are highly sought after, only one enzyme that can convert fluoride to organic fluorine has been described8. Streptomyces cattleya can form carbon–fluorine bonds9 and must therefore have evolved an enzyme able to overcome the chemical challenges of using aqueous fluoride. Here we report the sequence and three-dimensional structure of the first native fluorination enzyme, 5′-fluoro-5′-deoxyadenosine synthase, from this organism. Both substrate and products have been observed bound to the enzyme, enabling us to propose a nucleophilic substitution mechanism for this biological fluorination reaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
O'Hagan, D. & Harper, D. B. Fluorine-containing natural products. J. Chem. 100, 127–133 (1999)
Xu, X.-H. et al. 5-Fluorouracil derivatives from the sponge Phakellia fusca. J. Nat. Prod. 66, 285–288 (2003)
vanPee, K. H. Biosynthesis of halogenated metabolites by bacteria. Annu. Rev. Microbiol. 50, 375–399 (1996)
Littlechild, J. Haloperoxidases and their role in biotransformation reactions. Curr. Opin. Chem. Biol. 3, 28–34 (1999)
Sandford, G. Organofluorine chemistry. Phil. Trans. R. Soc. Lond. A 358, 455–471 (2000)
Mann, J. Modern methods for the introduction of fluorine into organic molecules—an approach to compounds with altered chemical and biological activities. Chem. Soc. Rev. 16, 381–436 (1987)
Hutchinson, J. & Sandford, G. Elemental fluorine in organic chemistry. Top. Curr. Chem. 193, 1–43 (1997)
Zechel, D. L. et al. Enzymatic synthesis of carbon–fluorine bonds. J. Am. Chem. Soc. 123, 4350–4351 (2001)
Sanada, M. et al. Biosynthesis of fluorothreonine and fluoroacetic acid by the thienamycin producer, Streptomyces cattleya. J. Antibiotics 141, 259–265 (1986)
O'Hagan, D., Schaffrath, C., Cobb, S. L., Hamilton, J. T. G. & Murphy, C. D. Enzyme catalysed organofluorine synthesis. Nature 416, 279 (2002)
Schaffrath, C., Deng, H. & O'Hagan, D. Isolation and characterisation of 5′-fluorodeoxyadenosine synthetase, a fluorination enzyme from Streptomyces cattleya. FEBS Lett. 547, 111–114 (2003)
Boutselakis, H. et al. E-MSD: the European Bioinformatics Institute Macromolecular Structure Database. Nucleic Acids Res. 31, 458–462 (2003)
Holm, L. & Sander, C. Protein folds and families: sequence and structure alignments. Nucleic Acids Res. 27, 244–247 (1999)
Bateman, A. et al. The Pfam Protein Families Database. Nucleic Acids Res. 30, 276–280 (2002)
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Allen, F. H. The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr. B 58, 380–388 (2002)
Berman, H. M. et al. The Protein Data Bank. Acta Crystallogr. D 58, 899–907 (2002)
Kollman, P. A. et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897 (2000)
O'Hagan, D. et. al. An assay for the enantiomeric assay of [2H1]-fluoroacetic acid: Insight into the stereochemical course of fluorination during fluorometabolite biosynthesis in Streptomyces cattleya. J. Am. Chem. Soc. 125, 379–387 (2003)
Dunitz, J. & Taylor, R. Organic fluorine hardly ever accepts hydrogen bonds. Chem. Eur. J. 3, 89–98 (1997)
Howard, J. A. K., Hoy, J. V., O'Hagan, D. & Smith, G. T. How good is fluorine as a hydrogen bond acceptor? Tetrahedron 38, 12613–12622 (1996)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. Model 14, 354–360 (1996)
Dong, C. et al. Crystallization and X-ray diffraction of the 5′-fluoro-5′-deoxyadenosine synthase, a fluorination enzyme from Streptomyces cattleya. Acta Crystallogr. D 60, 760–763 (2003)
Doublie, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997)
Bailey, S. The CCP4 Suite—programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Morris, R. J., Perrakis, A. & Lamzin, V. S. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D 58, 968–975 (2002)
Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55, 247–255 (1999)
van Aalten, D. M. F. et al. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 10, 255–262 (1996)
Schaffrath, C., Cobb, S. L. & O'Hagan, D. Cell-free biosynthesis of fluoroacetate and 4-fluorothreonine in Streptomyces cattleya. Angew. Chem. Int. Edn Engl. 41, 3913–3915 (2002)
DeLano, W. L., The PyMOL Molecular Graphics System 〈http://www.pymol.org/〉 (2003).
Acknowledgements
We thank M. Dorward for technical assistance and G. Leonard for help with data collection. J.H.N. is a Biotechnology and Biological Sciences Research Council (BBSRC) Career Development Fellow; H.D. was supported by the BBSRC; D.O.H. thanks the BBSRC for financial support; J.H.N. thanks the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Dong, C., Huang, F., Deng, H. et al. Crystal structure and mechanism of a bacterial fluorinating enzyme. Nature 427, 561–565 (2004). https://doi.org/10.1038/nature02280
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02280
This article is cited by
-
Engineering non-haem iron enzymes for enantioselective C(sp3)–F bond formation via radical fluorine transfer
Nature Synthesis (2024)
-
Whole-cell catalysis by surface display of fluorinase on Escherichia coli using N-terminal domain of ice nucleation protein
Microbial Cell Factories (2021)
-
Enzymatic synthesis of fluorinated compounds
Applied Microbiology and Biotechnology (2021)
-
A fluoride-responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida
Nature Communications (2020)
-
Functional characterisation of the transcriptome from leaf tissue of the fluoroacetate-producing plant, Dichapetalum cymosum, in response to mechanical wounding
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.