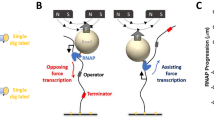
Abstract
Escherichia coli RNA polymerase (RNAP) synthesizes RNA with remarkable fidelity in vivo1. Its low error rate may be achieved by means of a ‘proofreading’ mechanism comprised of two sequential events. The first event (backtracking) involves a transcriptionally upstream motion of RNAP through several base pairs, which carries the 3′ end of the nascent RNA transcript away from the enzyme active site. The second event (endonucleolytic cleavage) occurs after a variable delay and results in the scission and release of the most recently incorporated ribonucleotides, freeing up the active site. Here, by combining ultrastable optical trapping apparatus with a novel two-bead assay to monitor transcriptional elongation with near-base-pair precision, we observed backtracking and recovery by single molecules of RNAP. Backtracking events (∼5 bp) occurred infrequently at locations throughout the DNA template and were associated with pauses lasting 20 s to >30 min. Inosine triphosphate increased the frequency of backtracking pauses, whereas the accessory proteins GreA and GreB, which stimulate the cleavage of nascent RNA, decreased the duration of such pauses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Erie, D. A., Yager, T. D. & von Hippel, P. H. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annu. Rev. Biophys. Biomol. Struct. 21, 379–415 (1992)
Jeon, C. & Agarwal, K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc. Natl Acad. Sci. USA 93, 13677–13682 (1996)
Thomas, M. J., Platas, A. A. & Hawley, D. K. Transcriptional fidelity and proofreading by RNA polymerase II. Cell 93, 627–637 (1998)
Erie, D. A., Hajiseyedjavadi, O., Young, M. C. & von Hippel, P. H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science 262, 867–873 (1993)
Kunkel, T. A. & Bebenek, K. DNA replication fidelity. Annu. Rev. Biochem. 69, 497–529 (2000)
Komissarova, N. & Kashlev, M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 272, 15329–15338 (1997)
Nudler, E., Mustaev, A., Lukhtanov, E. & Goldfarb, A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89, 33–41 (1997)
Marr, M. T. & Roberts, J. W. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell 6, 1275–1285 (2000)
Komissarova, N. & Kashlev, M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA 94, 1755–1760 (1997)
Reeder, T. C. & Hawley, D. K. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell 87, 767–777 (1996)
Tornaletti, S., Reines, D. & Hanawalt, P. C. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J. Biol. Chem. 274, 24124–24130 (1999)
Neuman, K. C., Abbondanzieri, E. A., Landick, R., Gelles, J. & Block, S. M. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell 115, 437–447 (2003)
Forde, N. R., Izhaky, D., Woodcock, G. R., Wuite, G. J. & Bustamante, C. Using mechanical force to probe the mechanism of pausing and arrest during continuous elongation by Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA 99, 11682–11687 (2002)
Schafer, D. A., Gelles, J., Sheetz, M. P. & Landick, R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature 352, 444–448 (1991)
Wang, M. D. et al. Force and velocity measured for single molecules of RNA polymerase. Science 282, 902–907 (1998)
Yin, H., Landick, R. & Gelles, J. Tethered particle motion method for studying transcript elongation by a single RNA polymerase molecule. Biophys. J. 67, 2468–2478 (1994)
Adelman, K. et al. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc. Natl Acad. Sci. USA 99, 13538–13543 (2002)
Veigel, C. et al. The motor protein myosin-I produces its working stroke in two steps. Nature 398, 530–533 (1999)
deCastro, M. J., Fondecave, R. M., Clarke, L. A., Schmidt, C. F. & Stewart, R. J. Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nature Cell Biol. 2, 724–729 (2000)
Nishiyama, M., Muto, E., Inoue, Y., Yanagida, T. & Higuchi, H. Substeps within the 8-nm step of the ATPase cycle of single kinesin molecules. Nature Cell Biol. 3, 425–428 (2001)
Aboul-ela, F., Koh, D., Tinoco, I. Jr & Martin, F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 13, 4811–4824 (1985)
Martin, F. H., Castro, M. M., Aboul-ela, F. & Tinoco, I. Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 13, 8927–8938 (1985)
Borukhov, S., Sagitov, V. & Goldfarb, A. Transcript cleavage factors from E. coli. Cell 72, 459–466 (1993)
Yildiz, A. et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 (2003)
Feng, G. H., Lee, D. N., Wang, D., Chan, C. L. & Landick, R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J. Biol. Chem. 269, 22282–22294 (1994)
Lang, M. J., Asbury, C. L., Shaevitz, J. W. & Block, S. M. An automated two-dimensional optical force clamp for single molecule studies. Biophys. J. 83, 491–501 (2002)
Svoboda, K. & Block, S. M. Biological applications of optical forces. Annu. Rev. Biophys. Biomol. Struct. 23, 247–285 (1994)
Wang, M. D., Yin, H., Landick, R., Gelles, J. & Block, S. M. Stretching DNA with optical tweezers. Biophys. J. 72, 1335–1346 (1997)
Acknowledgements
We acknowledge intellectual contributions from J. Gelles, and we thank the entire Block Laboratory, especially K. Neuman, for support and discussions. We also thank A. Meyer for reading of the original manuscript. This work was supported by grants from the NIGMS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Shaevitz, J., Abbondanzieri, E., Landick, R. et al. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 426, 684–687 (2003). https://doi.org/10.1038/nature02191
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02191
This article is cited by
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)
-
Modulating mechanical stability of heterodimerization between engineered orthogonal helical domains
Nature Communications (2020)
-
The dynamic landscape of transcription initiation in yeast mitochondria
Nature Communications (2020)
-
Polarization induced control of optical trap potentials in binary liquids
Scientific Reports (2019)
-
Pause sequences facilitate entry into long-lived paused states by reducing RNA polymerase transcription rates
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.