Abstract
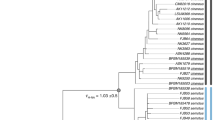
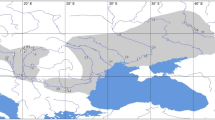
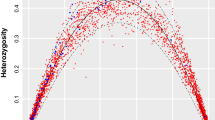
Examining patterns of inter-population genetic diversity can provide valuable information about both historical and current evolutionary processes affecting a species. Population genetic studies of flying and migratory species such as bats and birds have traditionally shown minimal population substructure, characterized by high levels of gene flow between populations1,2. In general, strongly substructured mammalian populations either are separated by non-traversable barriers or belong to terrestrial species with low dispersal abilities3. Species with female philopatry (the tendency to remain in or consistently return to the natal territory) might show strong substructure when examined with maternally inherited mitochondrial DNA, but this substructure generally disappears when biparentally inherited markers are used, owing to male-mediated gene flow4. Male-biased dispersal is considered typical for mammals5, and philopatry in both sexes is rare. Here we show strong population substructure in a migratory bat species, and philopatry in both sexes, as indicated by concordance of nuclear and mtDNA findings. Furthermore, the genetic structure correlates with local biomes and differentiation in wing morphology. There is therefore a close correlation of genetic and morphological differentiation in sympatric subspecific populations of this mammalian species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
McCracken, G. F., McCracken, M. K. & Vawter, A. T. Genetic structure in migratory populations of the bat Tadarida brasiliensis mexicana. J. Mammal. 75, 500–510 (1994)
Ball, R. M. J., Freeman, S., James, F. C., Bermingham, E. & Avise, J. C. Phylogeographic population structure of Red-winged blackbirds assessed by mitochondrial DNA. Proc. Natl Acad. Sci. USA 85, 1558–1562 (1988)
Avise, J. C. et al. Intraspecific phylogeography: The mtDNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18, 489–522 (1987)
Petit, E. & Mayer, F. Male dispersal in the noctule bat (Nyctalus noctula): Where are the limits? Proc. R. Soc. Lond. B 266, 1717–1722 (1999)
Greenwood, P. J. Mating systems, philopatry and dispersal in birds and mammals. Anim. Behav. 28, 1140–1162 (1980)
van der Merwe, M. Aspects of the social behaviour of the Natal clinging bat, Miniopterus schreibersii natalensis (A. Smith, 1834). Mammalia 37, 379–389 (1973)
Paetkau, D., Waits, L. P., Clarkson, P. L., Craighead, L. & Strobeck, C. An empirical evaluation of genetic distance statistics using microsatellite data from bear (Ursidae) populations. Genetics 147, 1943–1957 (1997)
Norman, J. A., Moritz, C. & Limpus, C. J. Mitochondrial DNA control region polymorphisms: Genetic markers for ecological studies of marine turtles. Mol. Ecol. 3, 363–373 (1994)
Worthington Wilmer, J., Moritz, C., Hall, L. & Toop, J. Extreme population structuring in the threatened ghost bat, Macroderma gigas: Evidence from mitochondrial DNA. Proc. R. Soc. Lond. B 257, 193–198 (1994)
Cardinal, B. R. & Christidis, L. Mitochondrial DNA and morphology reveal three geographically distinct lineages of the large bentwing bat (Miniopterus schreibersii) in Australia. Aust. J. Zool. 48, 1–19 (2000)
Wilkinson, G. S. The social organization of the common vampire bat. II. Mating system, genetic structure and relatedness. Behav. Ecol. Sociobiol. 17, 123–134 (1985)
Kerth, G., Mayer, F. & Konig, B. Mitochondrial DNA (mtDNA) reveals that female Bechstein's bats live in closed societies. Mol. Ecol. 9, 793–800 (2000)
Wilkinson, G. S. & Chapman, A. M. Length and sequence variation in evening bat D-Loop mtDNA. Genetics 128, 607–617 (1991)
Thompson, M. J. A. Roost philopatry in female pipistrelle bats Pipistrellus pipistrellus. J. Zool. 228, 673–679 (1992)
Amos, B., Schlotterer, C. & Tautz, D. Social structure of pilot whales revealed by analytical DNA profiling. Science 260, 670–672 (1993)
Burland, T. M., Barratt, E. M., Beaumont, M. A. & Racey, P. A. Population genetic structure and gene flow in a gleaning bat, Plecotus auritus. Proc. R. Soc. Lond. B 266, 975–980 (1999)
Palmeirim, J. M. & Rodrigues, L. in Ecology, Evolution and Behaviour of Bats (eds Racey, P. A. & Swift, S. M.) 219–231 (Clarendon, Oxford, 1995)
Norberg, U. M. & Rayner, J. M. V. Ecological morphology and flight in bats (Mammalia; Chiroptera): Wing adaptations, flight performance, foraging strategy and echolocation. Phil. Trans. R. Soc. Lond. B 316, 335–427 (1987)
Fenton, M. B. The foraging behaviour and ecology of animal-eating bats. Can. J. Zool. 68, 411–422 (1990)
Cowling, R. M., Richardson, D. M. & Pierce, S. M. (eds) Vegetation of Southern Africa (Cambridge Univ. Press, 1997)
Rautenbach, I. L., Kemp, A. C. & Scholtz, C. H. Fluctuations in availability of arthropods correlated with microchiropteran and avian predator activities. Koedoe 31, 77–90 (1988)
Bernard, R. T. F. Reproductive synchrony and annual variation in foetal growth rate in the long-fingered bat (Miniopterus schreibersii). J. Zool. 232, 485–490 (1994)
Racey, P. A. & Entwistle, A. C. in Reproductive Biology of Bats (eds Crichton, E. G. & Krutzsch, P. H.) 363–414 (Academic, London, 2000)
Worthington Wilmer, J. & Barratt, E. M. A non-lethal method of tissue sampling for genetic studies of Chiropterans. Bat Res. News 37, 1–3 (1996)
Miller-Butterworth, C. M., Jacobs, D. S. & Harley, E. H. Isolation and characterisation of highly polymorphic microsatellite loci in Schreibers' long-fingered bat, Miniopterus schreibersii (Chiroptera: Vespertilionidae). Mol. Ecol. Notes 2, 139–141 (2002)
Moore, S. S., Hale, P. & Byrne, K. NCAM: a polymorphic microsatellite locus conserved across eutherian mammal species. Anim. Genet. 29, 33–36 (1998)
Raymond, M., Rousset, F. et al. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 86, 248 (1995)
Goodman, S. RST Calc: a collection of computer programs for calculating unbiased estimates of genetic differentiation and determining their significance for microsatellite data. Mol. Ecol. 6, 881–885 (1997)
Saunders, M. B. & Barclay, R. M. Ecomorphology of insectivorous bats: A test of predictions using two morphologically similar species. Ecology 73, 1335–1345 (1992)
Harley, E. H. & Miller-Butterworth, C. M. A software assistant for bat wing measurements. Bat Res. News 41, 99–102 (2000)
Acknowledgements
We thank R. Bernard, P. Taylor, B. Eick, C. Logan, M. Kirkman and R. Louw for assistance with obtaining samples; P. Owen, I. Baumgarten, C. van Heerden and D. James for technical assistance in the laboratory; and M. Butterworth and E. Teeling for constructive comments on the manuscript. This work was funded by grants from the National Research Foundation of South Africa and the University Research Committee of the University of Cape Town.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Miller-Butterworth, C., Jacobs, D. & Harley, E. Strong population substructure is correlated with morphology and ecology in a migratory bat. Nature 424, 187–191 (2003). https://doi.org/10.1038/nature01742
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01742
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.