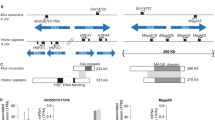
Abstract
Eight palindromes comprise one-quarter of the euchromatic DNA of the male-specific region of the human Y chromosome, the MSY1. They contain many testis-specific genes and typically exhibit 99.97% intra-palindromic (arm-to-arm) sequence identity1. This high degree of identity could be interpreted as evidence that the palindromes arose through duplication events that occurred about 100,000 years ago. Using comparative sequencing in great apes, we demonstrate here that at least six of these MSY palindromes predate the divergence of the human and chimpanzee lineages, which occurred about 5 million years ago. The arms of these palindromes must have subsequently engaged in gene conversion, driving the paired arms to evolve in concert. Indeed, analysis of MSY palindrome sequence variation in existing human populations provides evidence of recurrent arm-to-arm gene conversion in our species. We conclude that during recent evolution, an average of approximately 600 nucleotides per newborn male have undergone Y–Y gene conversion, which has had an important role in the evolution of multi-copy testis gene families in the MSY.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003)
Kuroda-Kawaguchi, T. et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genet. 29, 279–286 (2001)
Agulnik, A. I. et al. Evolution of the DAZ gene family suggests that Y-linked DAZ plays little, or a limited, role in spermatogenesis but underlines a recent African origin for human populations. Hum. Mol. Genet. 7, 1371–1377 (1998)
Reijo, R. et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nature Genet. 10, 383–393 (1995)
Glaser, B. et al. Simian Y chromosomes: species-specific rearrangements of DAZ, RBM, and TSPY versus contiguity of PAR and SRY. Mamm. Genome 9, 226–231 (1998)
Makova, K. D. & Li, W. H. Strong male-driven evolution of DNA sequences in humans and apes. Nature 416, 624–626 (2002)
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J. & Stahl, F. W. The double-strand-break repair model for recombination. Cell 33, 25–35 (1983)
Jackson, J. A. & Fink, G. R. Gene conversion between duplicated genetic elements in yeast. Nature 292, 306–311 (1981)
Underhill, P. A. et al. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 65, 43–62 (2001)
Murti, J. R., Bumbulis, M. & Schimenti, J. C. High-frequency germ line gene conversion in transgenic mice. Mol. Cell. Biol. 12, 2545–2552 (1992)
Johnson, R. D. & Jasin, M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407 (2000)
Bailey, J. A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002)
International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001)
Small, K., Iber, J. & Warren, S. T. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nature Genet. 16, 95–99 (1997)
Aradhya, S. et al. Multiple pathogenic and benign genomic rearrangements occur at a 35 kb duplication involving the NEMO and LAGE2 genes. Hum. Mol. Genet. 10, 2557–2567 (2001)
Rochette, C. F., Gilbert, N. & Simard, L. R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 108, 255–266 (2001)
Deeb, S. S., Jorgensen, A. L., Battisti, L., Iwasaki, L. & Motulsky, A. G. Sequence divergence of the red and green visual pigments in great apes and humans. Proc. Natl Acad. Sci. USA 91, 7262–7266 (1994)
Zhou, Y.-H. & Li, W.-H. Gene conversion and natural selection in the evolution of X-linked color vision genes in higher primates. Mol. Biol. Evol. 18, 780–783 (1996)
Charlesworth, B. & Charlesworth, D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. Lond. B 355, 1563–1572 (2000)
Bohossian, H. B., Skaletsky, H. & Page, D. C. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406, 622–625 (2000)
Kumar, S. & Hedges, S. B. A molecular timescale for vertebrate evolution. Nature 392, 917–920 (1998)
Fujiyama, A. et al. Construction and analysis of a human-chimpanzee comparative clone map. Science 295, 131–134 (2002)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Saxena, R. et al. Four DAZ genes in two clusters found in AZFc region of human Y chromosome. Genomics 67, 256–267 (2000)
Ohta, T. Allelic and nonallelic homology of a supergene family. Proc. Natl Acad. Sci. USA 79, 3251–3254 (1982)
Casanova, M. et al. A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 230, 1403–1406 (1985)
Underhill, P. A. et al. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 7, 996–1005 (1997)
Shen, P. et al. Population genetic implications from sequence variation in four Y chromosome genes. Proc. Natl Acad. Sci. USA 97, 7354–7359 (2000)
Acknowledgements
We thank R. K. Alagappan and L. G. Brown for technical contributions; N. A. Ellis, M. F. Hammer, T. Jenkins and P. A. Underhill for assistance with genealogical studies; H. M. McClure and Yerkes Regional Primate Research Center for samples; C. Disteche, A. E. Donnenfeld, J. H. Hersh, T. Jenkins, P. G. McDonough, B. McGillivray, R. D. Oates, P. Patrizio, R. Rosenfield, L. Shapiro, S. Silber, M. C. Summers, J. Weissenbach, B. Whitmire and S. Yang for patient samples; and J. E. Alfoldi, B. Charlesworth, A. G. Clark, J. Koubova, J. Lange, B. Levy, T. L. Orr-Weaver, S. Repping, W. R. Rice and J. Saionz for comments on the manuscript. This work was supported by the National Institutes of Health and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Rozen, S., Skaletsky, H., Marszalek, J. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003). https://doi.org/10.1038/nature01723
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature01723
This article is cited by
-
High incidence of AZF duplications in clan-structured Iranian populations detected through Y chromosome sequencing read depth analysis
Scientific Reports (2023)
-
Characterizing the evolution and phenotypic impact of ampliconic Y chromosome regions
Nature Communications (2023)
-
Dynamic alternative DNA structures in biology and disease
Nature Reviews Genetics (2023)
-
Eighty million years of rapid evolution of the primate Y chromosome
Nature Ecology & Evolution (2023)
-
Protein innovation through template switching in the Saccharomyces cerevisiae lineage
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.