Abstract
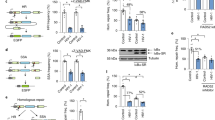
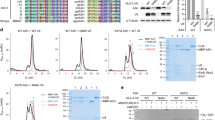
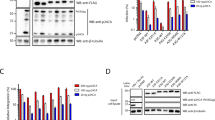
Viral replication usually requires that innate intracellular lines of defence be overcome, a task usually accomplished by specialized viral gene products. The virion infectivity factor (Vif) protein of human immunodeficiency virus (HIV) is required during the late stages of viral production to counter the antiviral activity of APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G; also known as CEM15), a protein expressed notably in human T lymphocytes1,2,3,4. When produced in the presence of APOBEC3G, vif-defective virus is non-infectious. APOBEC3G is closely related to APOBEC1, the central component of an RNA-editing complex that deaminates a cytosine residue in apoB messenger RNA5,6,7. APOBEC family members also have potent DNA mutator activity through dC deamination8; however, whether the editing potential of APOBEC3G has any relevance to HIV inhibition is unknown. Here, we demonstrate that it does, as APOBEC3G exerts its antiviral effect during reverse transcription to trigger G-to-A hypermutation in the nascent retroviral DNA. We also find that APOBEC3G can act on a broad range of retroviruses in addition to HIV, suggesting that hypermutation by editing is a general innate defence mechanism against this important group of pathogens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
von Schwedler, U., Song, J., Aiken, C. & Trono, D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67, 4945–4955 (1993)
Gabuzda, D. H. et al. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66, 6489–6495 (1992)
Simon, J. H. & Malim, M. H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70, 5297–5305 (1996)
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002)
Jarmuz, A. et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79, 285–296 (2002)
Powell, L. M. et al. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell 50, 831–840 (1987)
Teng, B., Burant, C. F. & Davidson, N. O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260, 1816–1819 (1993)
Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253 (2002)
MacGinnitie, A. J., Anant, S. & Davidson, N. O. Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity. J. Biol. Chem. 270, 14768–14775 (1995)
Zufferey, R., Nagy, D., Mandel, R. J., Naldini, L. & Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nature Biotechnol. 15, 871–875 (1997)
Kotsopoulou, E., Kim, V. N., Kingsman, A. J., Kingsman, S. M. & Mitrophanous, K. A. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 74, 4839–4852 (2000)
Negre, D. et al. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7, 1613–1623 (2000)
Mitrophanous, K. et al. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 6, 1808–1818 (1999)
Charneau, P. et al. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241, 651–662 (1994)
Jayanta, C., Ming, T., Chan, K., Eric, P. & Alt, F. W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730 (2003)
Klarmann, G. J., Chen, X., North, T. W. & Preston, B. D. Incorporation of uracil into minus strand DNA affects the specificity of plus strand synthesis initiation during lentiviral reverse transcription. J. Biol. Chem. 278, 7902–7909 (2003)
Borman, A. M., Quillent, C., Charneau, P., Kean, K. M. & Clavel, F. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G → A hypermutation during negative-strand viral DNA synthesis. Virology 208, 601–609 (1995)
Vartanian, J. P., Meyerhans, A., Asjo, B. & Wain-Hobson, S. Selection, recombination, and G → A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65, 1779–1788 (1991)
Wain-Hobson, S., Sonigo, P., Guyader, M., Gazit, A. & Henry, M. Erratic G → A hypermutation within a complete caprine arthritis-encephalitis virus (CAEV) provirus. Virology 209, 297–303 (1995)
Johnson, P. R., Hamm, T. E., Goldstein, S., Kitov, S. & Hirsch, V. M. The genetic fate of molecularly cloned simian immunodeficiency virus in experimentally infected macaques. Virology 185, 217–228 (1991)
Turelli, P., Guiguen, F., Mornex, J. F., Vigne, R. & Querat, G. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J. Virol. 71, 4522–4530 (1997)
Mansky, L. M. In vivo analysis of human T-cell leukemia virus type 1 reverse transcription accuracy. J. Virol. 74, 9525–9531 (2000)
Pathak, V. K. & Temin, H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl Acad. Sci. USA 87, 6019–6023 (1990)
Gunther, S. et al. Naturally occurring hepatitis B virus genomes bearing the hallmarks of retroviral G → A hypermutation. Virology 235, 104–108 (1997)
Turelli, P. et al. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 7, 1245–2154 (2001)
Dettenhofer, M. & Yu, X. F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73, 1460–1467 (1999)
Butler, S. L., Hansen, M. S. & Bushman, F. D. A quantitative assay for HIV DNA integration in vivo. Nature Med. 7, 631–634 (2001)
Yerly, S. et al. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354, 729–733 (1999)
Acknowledgements
We thank S. Vianin for technical assistance, and M. Malim, R. Mulligan, K. Mitrophanous, F.-L. Cosset and M. Matsuda for the gift of reagents. This work was supported by the Swiss National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Mangeat, B., Turelli, P., Caron, G. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003). https://doi.org/10.1038/nature01709
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature01709
This article is cited by
-
Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H
Nature Communications (2023)
-
Regulatory variants of APOBEC3 genes potentially associate with COVID-19 severity in populations with African ancestry
Scientific Reports (2023)
-
Antiretroviral APOBEC3 cytidine deaminases alter HIV-1 provirus integration site profiles
Nature Communications (2023)
-
ADAR1 is a promising risk stratification biomarker of remnant liver recurrence after hepatic metastasectomy for colorectal cancer
Scientific Reports (2023)
-
Distinct monkeypox virus lineages co-circulating in humans before 2022
Nature Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.