Abstract
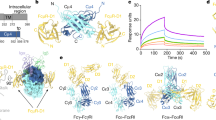
Immunoglobulin-α (IgA)-bound antigens induce immune effector responses by activating the IgA-specific receptor FcαRI (CD89) on immune cells. Here we present crystal structures of human FcαRI alone and in a complex with the Fc region of IgA1 (Fcα). FcαRI has two immunoglobulin-like domains that are oriented at approximately right angles to each other. Fcα resembles the Fcs of immunoglobulins IgG and IgE, but has differently located interchain disulphide bonds and external rather than interdomain N-linked carbohydrates. Unlike 1:1 FcγRIII:IgG and FcɛRI:IgE complexes, two FcαRI molecules bind each Fcα dimer, one at each Cα2–Cα3 junction. The FcαRI-binding site on IgA1 overlaps the reported polymeric immunoglobulin receptor (pIgR)-binding site, which might explain why secretory IgA cannot initiate phagocytosis or bind to FcαRI-expressing cells in the absence of an integrin co-receptor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Monteiro, R. C. & Van De Winkel, J. G. IgA Fc receptors. Annu. Rev. Immunol. 21, 177–204 (2003)
Brandtzaeg, P. et al. Regional specialization in the mucosal immune system: what happens in the microcompartments? Immunol. Today 20, 141–151 (1999)
Norderhaug, I. N., Johansen, F. E., Schjerven, H. & Brandtzaeg, P. Regulation of the formation and external transport of secretory immunoglobulins. Crit. Rev. Immunol. 19, 481–508 (1999)
Monteiro, R. C., Kubagawa, H. & Cooper, M. D. Cellular distribution, regulation, and biochemical nature of an Fcα receptor in humans. J. Exp. Med. 171, 597–613 (1990)
Vidarsson, G. et al. Activity of human IgG and IgA subclasses in immune defense against Neisseria meningitidis serogroup B. J. Immunol. 166, 6250–6256 (2001)
van Egmond, M. et al. FcαRI-positive liver Kupffer cells: reappraisal of the function of immunoglobulin A in immunity. Nature Med. 6, 680–685 (2000)
van Spriel, A. B. et al. Mac-1 (CD11b/CD18) is essential for Fc receptor-mediated neutrophil cytotoxicity and immunologic synapse formation. Blood 97, 2478–2486 (2001)
Motegi, Y. & Kita, H. Interaction with secretory component stimulates effector functions of human eosinophils but not of neutrophils. J. Immunol. 161, 4340–4346 (1998)
Wende, H., Colonna, M., Ziegler, A. & Volz, A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm. Genome 10, 154–160 (1999)
Garman, S. C., Wurzburg, B. A., Tarchevskaya, S. S., Kinet, J. P. & Jardetzky, T. S. Structure of the Fc fragment of human IgE bound to its high-affinity receptor FcɛRIα. Nature 406, 259–266 (2000)
Sondermann, P., Huber, R., Oosthuizen, V. & Jacob, U. The 3.2-Å crystal structure of the human IgG1 Fc fragment-FcγRIII complex. Nature 406, 267–273 (2000)
Herr, A. B., White, C. L., Milburn, C., Wu, C. & Bjorkman, P. J. Bivalent binding of IgA1 to FcαRI suggests a mechanism for cytokine activation of IgA phagocytosis. J. Mol. Biol. 327, 645–657 (2003)
Pleass, R. J., Dunlop, J. I., Anderson, C. M. & Woof, J. M. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human Fcα receptor (FcαR) CD89. J. Biol. Chem. 274, 23508–23514 (1999)
Carayannopoulos, L., Hexham, J. M. & Capra, J. D. Localization of the binding site for the monocyte immunoglobulin (Ig) A-Fc receptor (CD89) to the domain boundary between Cα2 and Cα3 in human IgA1. J. Exp. Med. 183, 1579–1586 (1996)
Martin, W. L., West, A. P. Jr, Gan, L. & Bjorkman, P. J. Crystal structure at 2.8 Å of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol. Cell 7, 867–877 (2001)
Chapman, T. L., Heikema, A. P., West, A. P. & Bjorkman, P. J. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2). Immunity 13, 727–736 (2000)
Garman, S. C., Kinet, J. P. & Jardetzky, T. S. Crystal structure of the human high-affinity IgE receptor. Cell 95, 951–961 (1998)
Maxwell, K. F. et al. Crystal structure of the human leukocyte Fc receptor, FcγRIIa. Nature Struct. Biol. 6, 437–442 (1999)
Sondermann, P., Huber, R. & Jacob, U. Crystal structure of the soluble form of the human Fcγ-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 Å resolution. EMBO J. 18, 1095–1103 (1999)
Fan, Q. R. et al. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature 389, 96–100 (1997)
Huber, R., Deisenhofer, J., Colman, P. M., Matsushima, M. & Palm, W. Crystallographic structure studies of an IgG molecule and an Fc fragment. Nature 264, 415–420 (1976)
Wan, T. et al. The crystal structure of IgE Fc reveals an asymmetrically bent conformation. Nature Immunol. 3, 681–686 (2002)
Wurzburg, B. A., Garman, S. C. & Jardetzky, T. S. Structure of the human IgE-Fc Cɛ3-Cɛ4 reveals conformational flexibility in the antibody effector domains. Immunity 13, 375–385 (2000)
Putnam, F. W., Liu, Y. S. & Low, T. L. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the α1 heavy chain. J. Biol. Chem. 254, 2865–2874 (1979)
Yang, C., Kratzin, H., Gotz, H. & Hilschmann, N. Rule of antibody structure. Primary structure of a human monoclonal IgA-immunoglobulin (myeloma protein Tro). VII. Purification and characterization of the disulfide bridges. Hoppe-Seyler's Z. Physiol. Chem. 360, 1919–1940 (1979)
Morton, H. C. et al. Immunoglobulin-binding sites of human FcαRI (CD89) and bovine Fcγ2R are located in their membrane-distal extracellular domains. J. Exp. Med. 189, 1715–1722 (1999)
Wines, B. D. et al. Identification of residues in the first domain of human Fcα receptor essential for interaction with IgA. J. Immunol. 162, 2146–2153 (1999)
Wines, B. D., Sardjono, C. T., Trist, H. M., Lay, C. S. & Hogarth, P. M. The interaction of FcαRI with IgA and its implications for ligand binding by immunoreceptors of the leukocyte receptor cluster. J. Immunol. 166, 1781–1789 (2001)
Garman, S. C., Sechi, S., Kinet, J. P. & Jardetzky, T. S. The analysis of the human high affinity IgE receptor FcɛRIα from multiple crystal forms. J. Mol. Biol. 311, 1049–1062 (2001)
Deisenhofer, J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry 20, 2361–2370 (1981)
Sauer-Eriksson, A. E., Kleywegt, G. J., Uhlen, M. & Jones, T. A. Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure 3, 265–278 (1995)
Corper, A. L. et al. Structure of human IgM rheumatoid factor Fab bound to its autoantigen IgG Fc reveals a novel topology of antibody–antigen interaction. Nature Struct. Biol. 4, 374–381 (1997)
DeLano, W. L., Ultsch, M. H., de Vos, A. M. & Wells, J. A. Convergent solutions to binding at a protein–protein interface. Science 287, 1279–1283 (2000)
Saphire, E. O. et al. Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 319, 9–18 (2002)
Shogren, R., Gerken, T. A. & Jentoft, N. Role of glycosylation on the conformation and chain dimensions of O-linked glycoproteins: light-scattering studies of ovine submaxillary mucin. Biochemistry 28, 5525–5536 (1989)
Iikura, M. et al. Secretory IgA induces degranulation of IL-3-primed basophils. J. Immunol. 161, 1510–1515 (1998)
Hexham, J. M. et al. A human immunoglobulin (Ig)A Cα3 domain motif directs polymeric Ig receptor-mediated secretion. J. Exp. Med. 189, 747–752 (1999)
White, K. D. & Capra, J. D. Targeting mucosal sites by polymeric immunoglobulin receptor-directed peptides. J. Exp. Med. 196, 551–555 (2002)
Bracke, M., Lammers, J. W., Coffer, P. J. & Koenderman, L. Cytokine-induced inside-out activation of FcαR (CD89) is mediated by a single serine residue (S263) in the intracellular domain of the receptor. Blood 97, 3478–3483 (2001)
Weisbart, R. H., Kacena, A., Schuh, A. & Golde, D. W. GM-CSF induces human neutrophil IgA-mediated phagocytosis by an IgA Fc receptor activation mechanism. Nature 332, 647–648 (1988)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
De La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997)
Collaborative Computational Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994)
McDonald, I. K. & Thornton, J. M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 (1994)
Su, X. D. et al. Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion. Science 281, 991–995 (1998)
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Merritt, E. A. & Murphy, M. E. P. Raster3D Version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994)
Acknowledgements
We thank E. R. Sprague for help with data collection; E. R. Sprague, L. M. Thomas, A. P. West and K. Locher for discussions; P. M. Snow and the Caltech Protein Expression Facility for FcαRI expression; C. L. White for initial FcαRI crystallization trials; staff of the Stanford Synchrotron Radiation Laboratory for technical support; and G. Waksman, M. J. Bennett, W. L. Martin and members of the Bjorkman laboratory for comments on the manuscript. This work was supported by funds from the Damon Runyon Cancer Research Foundation (to A.B.H.), the Howard Hughes Medical Institute (to P.J.B.) and the Ralph M. Parsons Foundation for computational support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
41586_2003_BFnature01685_MOESM1_ESM.doc
Supplementary Tables: Table 1: Crystallographic statistics Table 2: RMS deviations between FcαRI or Fcα and related proteinsTable 3: Pairwise interactions in the FcαRI:Fcα interface (DOC 131 kb)
Rights and permissions
About this article
Cite this article
Herr, A., Ballister, E. & Bjorkman, P. Insights into IgA-mediated immune responses from the crystal structures of human FcαRI and its complex with IgA1-Fc. Nature 423, 614–620 (2003). https://doi.org/10.1038/nature01685
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature01685
This article is cited by
-
SIgA structures bound to Streptococcus pyogenes M4 and human CD89 provide insights into host-pathogen interactions
Nature Communications (2023)
-
Variable-heavy (VH) families influencing IgA1&2 engagement to the antigen, FcαRI and superantigen proteins G, A, and L
Scientific Reports (2022)
-
Secret(ory) revealed: the long-awaited structures of secretory IgA
Cell Research (2020)
-
Igh locus structure and evolution in Platyrrhines: new insights from a genomic perspective
Immunogenetics (2020)
-
Role of IgA receptors in the pathogenesis of IgA nephropathy
Journal of Nephrology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.