Abstract
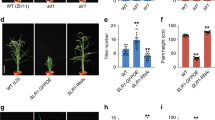
Tillering in rice (Oryza sativa L.) is an important agronomic trait for grain production, and also a model system for the study of branching in monocotyledonous plants. Rice tiller is a specialized grain-bearing branch that is formed on the unelongated basal internode and grows independently of the mother stem (culm) by means of its own adventitious roots1. Rice tillering occurs in a two-stage process: the formation of an axillary bud at each leaf axil and its subsequent outgrowth2. Although the morphology and histology2,3 and some mutants of rice tillering4 have been well described, the molecular mechanism of rice tillering remains to be elucidated. Here we report the isolation and characterization of MONOCULM 1 (MOC1), a gene that is important in the control of rice tillering. The moc1 mutant plants have only a main culm without any tillers owing to a defect in the formation of tiller buds. MOC1 encodes a putative GRAS family nuclear protein that is expressed mainly in the axillary buds and functions to initiate axillary buds and to promote their outgrowth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Li, Y.-H. Morphology and Anatomy of Grass Family Crops 138–142 (Shanghai Science and Technology Press, Shanghai, 1979)
Hanada, K. in Science of the Rice Plant Vol. 1 Morphology (eds Matsuo, T. & Hoshikawa, K.) 222–258 (Food and Agriculture Policy Research Center, Tokyo, 1993)
Hanada, K. in Science of the Rice Plant Vol. 2 Physiology (eds Matsuo, T., Kumazawa, K., Ishii, R., Ishihara, K. & Hirata, H.) 61–65 (Food and Agriculture Policy Research Center, Tokyo, 1995)
Iwata, N., Takamure, I., Wu, H.-K., Siddinq, E. A. & Rutger, J. N. List of genes for various traits (with chromosome and main literature). Rice Genet. Newsl. 12, 61–93 (1995)
Schumacher, K., Schmitt, T., Rossberg, M., Schmitz, G. & Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl Acad. Sci. USA 96, 290–295 (1999)
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D. & Benfey, P. N. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119 (1999)
Di Laurenzio, L. et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433 (1996)
Helariutta, Y. et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000)
Bolle, C., Koncz, C. & Chua, N. H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14, 1269–1278 (2000)
Peng, J. et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205 (1997)
Silverstone, A. L., Ciampaglio, C. N. & Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169 (1998)
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M. & Matsuoka, M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70 (2002)
Peng, J. et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999)
Richards, D. E., Peng, J. & Harberd, N. P. Plant GRAS and metazoan STATs: one family? Bioessays 22, 573–577 (2000)
Smith, H. M., Hicks, G. R. & Raikhel, N. V. Importin α from Arabidopsis thaliana is a nuclear import receptor that recognizes three classes of import signals. Plant Physiol. 114, 411–417 (1997)
Stirnberg, P., van De Sande, K. & Leyser, H. M. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141 (2002)
Katayama, T. Analytical studies of tillering in paddy rice. J. Imp. Agric. Exp. Stn Jpn 1, 327–374 (1931)
Yan, J.-Q., Zhu, J., He, C.-X., Benmoussa, M. & Wu, P. Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.). Theor. Appl. Genet. 97, 267–274 (1998)
Sato, Y. et al. A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl Acad. Sci. USA 93, 8117–8122 (1996)
Doebley, J., Stec, A. & Hubbard, L. The evolution of apical dominance in maize. Nature 386, 485–488 (1997)
Lukens, L. & Doebley, J. Molecular evolution of the teosinte branched gene among maize and related grasses. Mol. Biol. Evol. 18, 627–638 (2001)
Hubbard, L., McSteen, P., Doebley, J. & Hake, S. Expression patterns and mutant phenotype of teosinte branched 1 correlate with growth suppression in maize and teosinte. Genetics 162, 1927–1935 (2002)
Schmitz, G. et al. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl Acad. Sci. USA 99, 1064–1069 (2002)
Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N. & Clark, S. E. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236 (2001)
Harushima, Y. et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494 (1998)
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994)
Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M. & Kobayashi, H. Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18, 455–463 (1999)
Fobert, P. R., Coen, E. S., Murphy, G. J. & Doonan, J. H. Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 13, 616–624 (1994)
Hu, Y.-X., Bao, F. & Li, J. Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24, 693–701 (2000)
Acknowledgements
We thank J. Zuo, N.-H. Chua and X.-W. Deng for critical comments on the manuscript, B. Zhang for assistance in using the microscope, I. Takamure for providing rcn1–rcn5 mutants, the MAFF DNA Bank of Japan for providing RFLP probes and YAC clones, the Clemson University Genomic Institute for providing Nipponbare BAC library filters and clones, Y. Niwa for providing the CaMV35SΩ-sGFP (S65T)-NOS-3′ construct, and M. Matsuoka for the OSH1 cDNA clone. F.H. is a visiting scientist from the Japan International Research Center for Agricultural Sciences, Tsukuba, Japan. This work was supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China and the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Li, X., Qian, Q., Fu, Z. et al. Control of tillering in rice. Nature 422, 618–621 (2003). https://doi.org/10.1038/nature01518
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01518
This article is cited by
-
GRAS gene family in rye (Secale cereale L.): genome-wide identification, phylogeny, evolutionary expansion and expression analyses
BMC Plant Biology (2024)
-
Wollastonite powder application increases rice yield and CO2 sequestration in a paddy field in Northeast China
Plant and Soil (2024)
-
Genetic and functional mechanisms of yield-related genes in rice
Acta Physiologiae Plantarum (2024)
-
Comparative transcriptome analysis and genetic dissection of vegetative branching traits in foxtail millet (Setaria italica)
Theoretical and Applied Genetics (2024)
-
Elevated CO2 Priming as a Sustainable Approach to Increasing Rice Tiller Number and Yield Potential
Rice (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.