Abstract
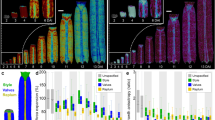
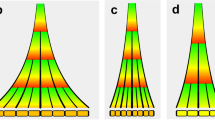
Development commonly involves the generation of complex shapes from simpler ones. One way of following this process is to use landmarks to track the fate of particular points in a developing organ1,2,3,4,5,6,7, but this is limited by the time over which it can be monitored. Here we use an alternative method, clonal analysis8, whereby dividing cells are genetically marked and their descendants identified visually, to observe the development of Antirrhinum (snapdragon) petals. Clonal analysis has previously been used to estimate growth parameters of leaves9,10,11 and Drosophila wings12,13,14 but these results were not integrated within a dynamic growth model. Here we develop such a model and use it to show that a key aspect of shape—petal asymmetry—in the petal lobe of Antirrhinum depends on the direction of growth rather than regional differences in growth rate. The direction of growth is maintained parallel to the proximodistal axis of the flower, irrespective of changes in shape, implying that long-range signals orient growth along the petal as a whole. Such signals may provide a general mechanism for orienting growth in other growing structures.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Avery, G. S. Structure and development of the tobacco leaf. Am. J. Bot. 20, 565–592 (1933)
Richards, O. W. & Kavanagh, A. J. The analysis of the relative growth gradients and changing form of growing organisms: Illustrated by the tobacco leaf. Am. Nat. 77, 385–399 (1943)
Erickson, R. O. Modeling of plant growth. Annu. Rev. Plant Physiol. 27, 407–434 (1976)
Erickson, R. O. Relative elemental rates and anisotropy of growth in area: a computer programme. J. Exp. Bot. 17, 390–403 (1966)
Wolf, S. D., Silk, W. K. & Plant, R. E. Quantitative patterns of leaf expansion—Comparison of normal and malformed leaf growth in Vitis-Vinifera Cv Ruby Red. Am. J. Bot. 73, 832–846 (June 1986)
Dumais, J. & Kwiatkowska, D. Analysis of surface growth in shoot apices. Plant J. 31, 229–241 (2002)
Hernandez, F., Havelange, A., Bernier, G. & Green, P. B. Growth behavior of single epidermal cells during flower formation: Sequential scanning electron micrographs provide kinematic patterns for Anagallis. Planta 185, 139–147 (1991)
Subtelny, S. & Sussex, I. M. (eds) The Clonal Basis of Development (Academic, New York, 1978)
Poethig, R. S. & Sussex, I. M. The cellular parameters of leaf development in tobacco: a clonal analysis. Planta 165, 170–184 (1985)
Poethig, R. S. & Szymkowiak, R. J. Clonal analysis of leaf development in maize. Maydica 40, 67–76 (1995)
Dolan, L. & Poethig, R. S. Clonal analysis of leaf development in cotton. Am. J. Bot. 85, 315–321 (1998)
Garcia-Bellido, A. & Merriam, J. R. Parameters of the wing imaginal disc development of Drosophila melanogaster. Dev. Biol. 24, 61–87 (1971)
Gonzalez-Gaitan, M., Capdevila, M. P. & Garcia-Bellido, A. Cell proliferation patterns in the wing imaginal disc of Drosophila. Mech. Dev. 40, 183–200 (1994)
Resino, J., Salama-Cohen, P. & Garcia-Bellido, A. Determining the role of patterned cell proliferation in the shape and size of the Drosophila wing. Proc Natl Acad. Sci. USA 99, 7502–7507 (2002)
Poethig, R. S. Clonal analysis of cell lineage patterns in plant development. Am. J. Bot. 74, 581–594 (1987)
Almeida, J., Rocheta, M. & Galego, L. Genetic control of flower shape in Antirrhinum majus. Development 124, 1387–1392 (1997)
Luo, D. et al. Control of organ asymmetry in flowers of Antirrhinum. Cell 99, 367–376 (1999)
Luo, D., Carpenter, R., Vincent, C., Copsey, L. & Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799 (1995)
Cubas, P., Lauter, N., Doebley, J. & Coen, E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222 (1999)
Vincent, C. A., Carpenter, C. & Coen, E. S. Plant J. 33, 1–10 (2003)
Perbal, M. C., Haughn, G., Saedler, H. & Schwarz-Sommer, Z. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122, 3433–3441 (1996)
Efremova, N. et al. Epidermal control of floral organ identity by class B homeotic genes in Antirrhinum and Arabidopsis. Development 14, 2661–2671 (2001)
Tilney-Basset, R. A. E. Plant Chimeras (Edward Arnolds, London, 1986)
Maksymowych, R. Analysis of Leaf Development Developmental and cell biology series 1 (Cambridge Univ. Press, London, 1973)
Lloyd, C. W. & Traas, J. A. The role of F-actin in determining the division plane of carrot suspension cells—drug studies. Development 102, 211–221 (1988)
Thompson, D. A. W. On Growth and Form, 2nd edn Vol. 2 (Cambridge Univ. Press, Cambridge, UK, 1942)
Sachs, T. in Pattern Formation in Plant Tissues (eds Barlow, P. W., Bray, D., Green, P. B. & Slack, J. M. W.) (Cambridge Univ. Press, Cambridge, UK, 1991)
Teleman, A., Strigini, M. & Cohen, S. M. Shaping morphogen gradients. Cell 105, 559–562 (2001)
Vincent, C. A., Carpenter, R. & Coen, E. S. Cell lineage patterns and homeotic gene activity during Antirrhinum flower development. Curr. Biol. 5, 1449–1457 (1995)
Harrison, B. J. & Fincham, J. R. S. Instability at the Pal locus in Antirrhinum Majus. I. Effects of environment on frequencies of somatic and germinal mutation. Heredity 19, 237–258 (1964)
Acknowledgements
We thank R. Carpenter for providing plant stocks, C. Vincent and K. Lee for the scanning electron microscopy, J. Dumais for help with SEM 3D reconstructions, N. Orme for drawings in Fig. 1, S. J. Impey for gathering the image database and for early work on the project, supported by a BBSRC grant. A.-G. R.-L. was supported by a Norwich Research Park studentship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Rolland-Lagan, AG., Bangham, J. & Coen, E. Growth dynamics underlying petal shape and asymmetry. Nature 422, 161–163 (2003). https://doi.org/10.1038/nature01443
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01443
This article is cited by
-
Molecular understanding of postharvest flower opening and senescence
Molecular Horticulture (2021)
-
Reduction in organ–organ friction is critical for corolla elongation in morning glory
Communications Biology (2021)
-
Real-time conversion of tissue-scale mechanical forces into an interdigitated growth pattern
Nature Plants (2021)
-
Molecular cloning and subcellular localization of six HDACs and their roles in response to salt and drought stress in kenaf (Hibiscus cannabinus L.)
Biological Research (2019)
-
Tissue cartography: compressing bio-image data by dimensional reduction
Nature Methods (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.