Abstract
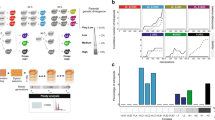
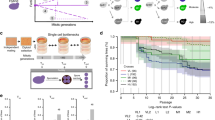
The Saccharomyces ‘sensu stricto’ yeasts are a group of species that will mate with one another, but interspecific pairings produce sterile hybrids. A retrospective analysis of their genomes revealed that translocations between the chromosomes of these species do not correlate with the group's sequence-based phylogeny1 (that is, translocations do not drive the process of speciation). However, that analysis was unable to infer what contribution such rearrangements make to reproductive isolation between these organisms. Here, we report experiments that take an interventionist, rather than a retrospective approach to studying speciation, by reconfiguring the Saccharomyces cerevisiae genome so that it is collinear with that of Saccharomyces mikatae. We demonstrate that this imposed genomic collinearity allows the generation of interspecific hybrids that produce a large proportion of spores that are viable, but extensively aneuploid. We obtained similar results in crosses between wild-type S. cerevisiae and the naturally collinear species Saccharomyces paradoxus, but not with non-collinear crosses. This controlled comparison of the effect of chromosomal translocation on species barriers suggests a mechanism for the generation of redundancy in the S. cerevisiae genome2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fischer, G. et al. Chromosomal evolution in Saccharomyces. Nature 405, 451–454 (2000)
Goffeau, A. et al. Life with 6000 genes. Science 274, 546–567 (1996)
Naumov, G. I. in The Expanding Realm of Yeast-like Fungi (eds de Hoog, G. S., Smith, M. T. & Weyman, A. C. M.) (Elsevier, Amsterdam, 1987)
Naumov, G. I., James, S. A., Naumova, E. S., Louis, E. J. & Roberts, I. N. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 50, 1931–1942 (2000)
Ryu, S. L., Murooka, Y. & Kaneko, Y. Reciprocal translocation at duplicated RPL2 loci might cause speciation of Saccharomyces bayanus and Saccharomyces cerevisiae. Curr. Genet. 33, 345–351 (1998)
Naumov, G. I., Naumova, E. S. & Querol, A. Genetic study of natural introgression supports delimitation of biological species in the Saccharomyces sensu stricto complex. System. Appl. Microbiol. 20, 595–601 (1997)
Marinoni, G. et al. Horizontal transfer of genetic material among Saccharomyces yeasts. J. Bacteriol. 181, 6488–6496 (1999)
Seoighe, C. & Wolfe, K. H. Yeast genome evolution in the post-genome era. Curr. Opin. Microbiol. 2, 548–554 (1999)
Delneri, D. et al. Exploring redundancy in the yeast genome: an improved strategy for the use of the cre-loxP system. Gene 252, 127–135 (2000)
Saccharomyces Genome Database. 〈http://genome-www4.stanford.edu/cgi-bin/FUNGI/FungiMap〉.
Delneri, D., Gardner, D. C. J. & Oliver, S. G. Analysis of the seven-member AAD gene set demonstrates that genetic redundancy in yeast may be more apparent than real. Genetics 153, 1591–1600 (1999)
Bengtsson, B. O. & Bodmer, W. F. The fitness of human translocation carriers. Ann. Human Genet. 40, 253–257 (1976)
Loidl, J., Jin, Q.-W. & Jantsch, M. Meiotic pairing and segregation of translocation quadrivalents in yeast. Chromosoma 107, 247–254 (1998)
Fischer, G., Neuvéglise, C., Durrens, P., Gaillardin, C. & Dujon, B. Evolution of gene order in the genomes of two related yeast species. Genome Res. 11, 2009–2019 (2001)
Lynch, M., O'Hely, M., Walsh, B. & Force, A. The probability of preservation of a newly arisen gene duplicate. Genetics 159, 1789–1804 (2001)
Ramsey, J. & Schemske, D. W. Pathways, mechanisms, and rate of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501 (1998)
Sebastiani, F., Barberio, C., Casalone, E., Cavalieri, D. & Polsinelli, M. Crosses between Saccharomyces cerevisiae and Saccharomyces bayanus generate fertile hybrids. Res. Microbiol. 153, 53–58 (2002)
Tamai, Y., Momma, T., Yoshimoto, H. & Kaneko, Y. Co-existence of two types of chromosome in the bottom-fermenting yeast, Saccharomyces pastorianus. Yeast 14, 923–937 (1998)
Kielland-Brandt, M. C., Nilson-Tillgren, T., Peterson, J. G. L., Holemberg, S. & Gjermansen, C. in Yeast Genetics (eds Spencer, J. F. T., Spencer, D. M. & Smith, A. R. W.) 421–438 (Springer, New York, 1983)
Wolfe, K. H. & Shields, D. C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713 (1997)
Llorente, B. et al. Genomic exploration of the hemiascomycetous yeasts: 20. Evolution of gene redundancy compared to Saccharomyces cerevisiae. FEBS Lett. 487, 122–133 (2000)
Greig, D., Borts, R. H., Louis, E. J. & Travisano, M. Epistasis and hybrid sterility in Saccharomyces. Proc. R. Soc. Lond. B. 269, 1167–1671 (2002)
Hunter, N., Chambers, S. R., Louis, E. J. & Borts, R. H. The mismatch repair system contributes to meiotic sterility in an interspecific yeast. EMBO J. 15, 1726–1733 (1996)
Adams, A., Gottschling, D. E., Kaiser, C. A. & Stearns, T. in Methods in Yeast Genetics (ed. Dickerson, M. M.) 145–175 (Cold Spring Harbor Laboratory Press, New York, 1997)
Gietz, R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360 (1995)
Gardner, D. C. J., Heale, S. M., Stateva, L. I. & Oliver, S. G. Treatment of yeast cells with wall lytic enzymes is not required to prepare chromosomes for pulsed-field gel analysis. Yeast 9, 1053–1055 (1993)
Acknowledgements
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (to E.J.L., I.N.R. and S.G.O.) and the Wellcome Trust (to S.G.O.). We thank S. James and L. Lockhart for their help in some early analyses, and B. Dujon for discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
41586_2003_BFnature01418_MOESM1_ESM.doc
Supplementary Information: Description of the strategy for engineering the translocations into S.cerevisiae, and two Tables listing all the oligonucleotides used in this work. (DOC 46 kb)
Rights and permissions
About this article
Cite this article
Delneri, D., Colson, I., Grammenoudi, S. et al. Engineering evolution to study speciation in yeasts. Nature 422, 68–72 (2003). https://doi.org/10.1038/nature01418
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01418
This article is cited by
-
Synthetic two-species allodiploid and three-species allotetraploid Saccharomyces hybrids with euploid (complete) parental subgenomes
Scientific Reports (2023)
-
Contribution of the SOS response and the DNA repair systems to norfloxacin induced mutations in E. coli
Marine Life Science & Technology (2023)
-
Macroevolutionary diversity of traits and genomes in the model yeast genus Saccharomyces
Nature Communications (2023)
-
Mitochondrial DNA duplication, recombination, and introgression during interspecific hybridization
Scientific Reports (2021)
-
Heterogeneous rates of genome rearrangement contributed to the disparity of species richness in Ascomycota
BMC Genomics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.