Abstract
The fundamental unit of eukaryotic chromatin, the nucleosome, consists of genomic DNA wrapped around the conserved histone proteins H3, H2B, H2A and H4, all of which are variously modified at their amino- and carboxy-terminal tails to influence the dynamics of chromatin structure and function1,2 — for example, conjugation of histone H2B with ubiquitin controls the outcome of methylation at a specific lysine residue (Lys 4) on histone H3, which regulates gene silencing in the yeast Saccharomyces cerevisiae3. Here we show that ubiquitination of H2B is also necessary for the methylation of Lys 79 in H3, the only modification known to occur away from the histone tails, but that not all methylated lysines in H3 are regulated by this 'trans-histone' pathway because the methylation of Lys 36 in H3 is unaffected. Given that gene silencing is regulated by the methylation of Lys 4 and Lys 79 in histone H3, we suggest that H2B ubiquitination acts as a master switch that controls the site-selective histone methylation patterns responsible for this silencing.
Similar content being viewed by others
Main
Lysine residues subject to methylation in yeast histone H3 are Lys 4 and Lys 36 near the amino terminus, and Lys 79 (refs 4–6, and data not shown), a modification site that is unique in that it is located away from the H3 tails in the first loop of the histone-fold domain. To identify what mediates methylation of Lys 79, we used an antibody raised against this methylated site (anti-H3 K79Me; Upstate Biotechnology) to screen nuclear extracts isolated from yeast strains containing deletions of known and putative histone methyltransferase enzymes.
We identified Dot1, a factor involved in gene silencing4,5, as a gene product that is essential for methylation of Lys 79 (Fig. 1a), in agreement with earlier findings4,5,6. This was unexpected, as Dot1 lacks the 'SET' domain, which until now was thought to be the only domain responsible for methylating histone lysine residues. No other protein containing the SET domain was found to mediate Lys-79 methylation (Fig. 1a). To confirm that Dot1 is the enzyme responsible, we showed that expression of DOT1 in a dot1-deleted strain restores Lys-79 methylation in H3 (see supplementary information) and that recombinant Dot1 contained methyltransferase activity towards nucleosomal H3 (Fig. 1b). Dot1, which resembles the arginine methyltransferase family in sequence and structure7, therefore represents a new class of lysine-specific histone methyltransferase enzymes.
a, Western blot of nuclear extracts isolated from mutant yeast strains carrying the indicated deletions and probed with antibodies against the methylated H3 lysine residues K79Me, K4Me or K36Me. b, Polyacrylamide-gel electrophoresis analysis of recombinant Dot1 for histone methyltransferase activity on nucleosomal substrates (Nuc) in vitro, as revealed by autoradiography (top) and Coomassie-blue staining of histones (bottom). c, Western blot of nuclear extracts from wild-type yeast and rad6-deleted or H2B K123R mutant strains probed with the histone antibodies indicated. Asterisks, an H3 proteolysis product that removes the methylation site at Lys 4 but not at Lys 36 or Lys 79.
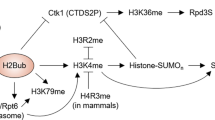
Ubiquitination of histone H2B is mediated by the enzyme Rad6, also known as ubiquitin-conjugating enzyme Ubc2 (ref. 8). We tested whether Rad6 ubiquitination of H2B could influence methylation at H3 sites apart from Lys 4 (ref. 3). Surprisingly, we found that there was a loss of Lys-79 methylation, but not of Lys-36 methylation, in strains deleted for RAD6 or mutated at the H2B-ubiquitination site (K123R; Fig. 1c). We verified that Dot1 is expressed in these mutant strains by using reverse transcription followed by polymerase chain reaction to detect the presence of its messenger RNA (see supplementary information). We conclude that Rad6 ubiquitination of H2B at Lys 123 specifically regulates the methylation of both Lys 4 and Lys 79 of H3 in a 'trans-histone' pathway.
We found that deletion of DOT1, SET1 (refs 9, 10) or SET2 (ref. 11) results in the specific loss of their respective modifications (Fig. 1a), indicating that the absence of these individual modifications does not affect the others. Given that the regulation of Lys-4 methylation by H2B ubiquitination is unidirectional3 and that Lys-4 methylation still occurs in the dot1-deleted (Fig. 1a) and Lys-79 mutant strains (see supplementary information), we conclude that the regulation of Lys-79 methylation by H2B ubiquitination is also unidirectional.
Our findings indicate that methylation of histone H3 at Lys 4 and Lys 79, but not at Lys 36, is regulated by the ubiquitination of histone H2B. As Lys 4 and Lys 79 methylation both mediate gene silencing, we propose that H2B ubiquitination acts as a master switch of gene silencing through a trans-histone pathway that leads to the appropriate patterns of histone methylation. Given that some lysine residues in H3 are affected by this pathway, but not others, our results lend support to the 'histone code' hypothesis2.
References
Kornberg, R. D. & Lorch, Y. Cell 98, 285–294 (1999).
Strahl, B. D. & Allis, C. D. Nature 403, 41–45 (2000).
Sun, Z. W. & Allis, C. D. Nature 418, 104–108 (2002).
van Leeuwen, F., Gafken, P. R. & Gottschling, D. E. Cell 109, 745–756 (2002).
Ng, H. H. et al. Genes Dev. 16, 1518–1527 (2002).
Feng, Q. et al. Curr. Biol. 12, 1052–1058 (2002).
Dlakic, M. Trends. Biochem. Sci. 26, 405–407 (2001).
Robzyk, K., Recht, J. & Osley, M. A. Science 287, 501–504 (2000).
Briggs, S. D. et al. Genes Dev. 15, 3286–3295 (2001).
Bryk, M. et al. Curr. Biol. 12, 165–170 (2002).
Strahl, B. D. et al. Mol. Cell Biol. 22, 1298–1306 (2002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
This advance online publication (AOP) Nature paper should be as “Author(s) Nature, 14 July 2002 (doi:10.1038/nature00970)”. Once the print version (identical to the AOP) is published, the citation becomes “Author(s) Nature volume, page (year); advance online publication 14 July 2002 (doi: 10.1038/nature00970)”.
Supplementary information
Rights and permissions
About this article
Cite this article
Briggs, S., Xiao, T., Sun, ZW. et al. Trans-histone regulatory pathway in chromatin. Nature 418, 498 (2002). https://doi.org/10.1038/nature00970
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature00970
This article is cited by
-
Context-defined cancer co-dependency mapping identifies a functional interplay between PRC2 and MLL-MEN1 complex in lymphoma
Nature Communications (2023)
-
CRISPR–ChIP reveals selective regulation of H3K79me2 by Menin in MLL leukemia
Nature Structural & Molecular Biology (2023)
-
H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity
Nature Chemical Biology (2022)
-
Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease
Nature Reviews Drug Discovery (2021)
-
Histone H2Bub1 deubiquitylation is essential for mouse development, but does not regulate global RNA polymerase II transcription
Cell Death & Differentiation (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.