Abstract
In recent years, messenger RNA (mRNA) has come into the spotlight as a versatile therapeutic with the potential to prevent and treat a staggering range of diseases. Billions of dollars have been invested in the commercial development of mRNA drugs, with ongoing clinical trials focused on vaccines (for example, influenza and Zika viruses) and cancer immunotherapy (for example, myeloma, leukaemia and glioblastoma). Although significant progress has been made in the design of in vitro-transcribed mRNA that retains potency while minimizing unwanted immune responses, the widespread use of mRNA drugs requires the development of safe and effective drug delivery vehicles. In this Review, we provide an overview of the field of mRNA therapeutics and describe recent advances in the development of synthetic materials that encapsulate and deliver mRNA payloads.
Similar content being viewed by others
Introduction
Messenger RNA (mRNA) is the middleman in the central dogma of molecular biology, which states that DNA is transcribed into mRNA, which is translated into protein. On the surface, the role of middleman is not a glamorous one: mRNA is neither a permanent genetic blueprint nor a functional end product. However, its transient nature confers tremendous flexibility and broader therapeutic utility than nearly all other classes of known drugs.
Ultimate control over disease requires the manipulation of protein expression. Unfortunately, the direct delivery of protein drugs is typically not practical because their large size and instability prevent them from reaching sufficiently high concentrations in vivo to induce a therapeutic effect1. Together with the cost of protein production, this limitation gave rise to the field of gene therapy, which focuses on the delivery of exogenously produced nucleic acids into cells. Although plasmid DNA was the first nucleic acid to be pursued as a therapeutic2, in vitro-transcribed (IVT) mRNA offers several advantages: it does not integrate into the genome, which is associated with a risk of carcinogenesis3, and a natural degradation pathway is in place to ensure that its activity is temporary. From a delivery perspective, mRNA, which lives in the cytoplasm, does not require transport across the formidable nuclear membrane4.
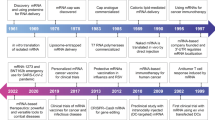
Discovered in 1961, 8 years after the DNA double-helix, mRNA was first delivered into cells by liposomes in the 1970s (Fig. 1). By the 1990s, delivery technology was sophisticated enough to support preclinical studies on mRNA-based vaccines and cancer immunotherapy5,6. Since then, mRNA therapeutics have shown considerable promise for both prophylaxis (for example, antiviral vaccines) and the treatment ofa broad range of diseases, including haemophilia B (Refs 7–9), myocardial infarction10, human immunodeficiency virus (HIV)11,12 and more than ten types of cancer13–15. These advances have been based on an improved pharmacological understanding of IVT mRNA as a drug, together with the development of delivery materials that are sufficiently efficacious to stimulate protein translation in the correct cell type.
Cas9, CRISPR-associated protein 9; DCs, dendritic cells; HIV, human immunodeficiency virus; iPSC, induced pluripotent stem cell; LNP, lipid nanoparticle; mRNA, messenger RNA; TALEN, transcription activator-like effector nuclease; ZFN, zinc finger nuclease.
Applications of mRNA drugs
In theory, mRNA drugs can prevent or treat any disease that is caused or affected by genetic components of the cell. Although, at first glance, it may seem as if mRNA therapy is appropriate only when gene upregulation is warranted, the advent of CRISPR technology has also enabled gene knockout upon delivery of mRNA encoding gene-editing nucleases. In the following section, we highlight the most promising applications of mRNA therapeutics.
Protein replacement therapy. In its most straightforward application, IVT mRNA is incorporated into the protein synthesis machinery of the target cell to induce expression of a desired protein (Fig. 2). Although some diseases (for example, haemophilia B (Ref. 16)) are characterized by insufficient protein expression and require protein augmentation, other diseases (for example, muscular dystrophy17) are caused by aberrant protein production that necessitates replacement18. Compared with the use of protein drugs for protein replacement, mRNA can be more effective on a per molecule basis, as a single mRNA molecule is translated into many copies of a protein over the course of hours or days19.
Upon entering the cell cytosol, in vitro-transcribed (IVT) mRNA is recognized by the translation machinery of the host, resulting in the binding of the ribosomal complex (step 1). The ribosomal complex scans the mRNA sequence in the 5′ to 3′ direction until it recognizes the start codon (initiation) and begins adding amino acids to the peptide chain with the aid of transfer RNAs (tRNAs) (elongation) (step 2). Elongation continues until the stop codon is encountered, prompting ribosomal release of the newly synthesized peptide (termination) (step 3). Finally, post-translational modifications convey functionality to the new protein (step 4).
In 1990, the first study to use IVT mRNA for protein production in vivo described the induction of luciferase expression following local injection of mRNA in mouse skeletal muscle20. Two years later, one of the first studies to treat a disease with an mRNA drug was reported21. Specifically, rats afflicted with diabetes insipidus were given intrahypothalamic injections of mRNA encoding the hormone vasopressin, which resulted in the reversal of disease symptoms within hours of treatment.
Importantly, protein replacement using IVT mRNA is feasible only when the mRNA is delivered into cells capable of folding the protein and inducing any required post-translational modifications. To attain full functionality, the majority of human proteins require post-translational modification, which occurs in the endoplasmic reticulum and Golgi apparatus22. Although there are hundreds of potential modifications, phosphorylation, acetylation and glycosylation are among the most common23. As such, protein replacement using IVT mRNA is ideal when transfecting cells that endogenously express the protein of interest and are therefore capable of appropriate modifications.
Vaccines. The most clinically advanced application of mRNA drugs — vaccines using antigen-encoding mRNA — are attractive alternatives to traditional vaccination strategies. Unlike live-attenuated viruses, which on rare occasions can revert to their pathogenic form, mRNA vaccines cannot replicate inside the body24. Furthermore, mRNA vaccines induce both cellular and humoral (antibody-based) immunity25,26 (Fig. 3).
After the transfection of antigen-encoding mRNA into the cytosol of an antigen-presenting cell (APC) (step 1), the mRNA is translated into antigenic peptide (step 2). This antigenic peptide is processed into smaller peptide epitopes (step 3) that bind to the major histocompatibility complex (MHC) class I (step 4a) or class II (step 4b). The MHCs are trafficked to the cell surface, where they present their antigenic epitopes to either CD8+ (cytotoxic) T cells (step 5a) or CD4+ (helper) T cells (step 5b), leading to cellular immunity or an antigen-specific antibody response, respectively25.
The development of antigen-specific immunity from an mRNA vaccine requires the transfection of antigen-presenting cells, such as dendritic cells. Administration is typically accomplished by intradermal, intramuscular or subcutaneous injection, as dendritic cells densely populate skin tissue27 and skeletal muscle28. Reports of mRNA-based vaccination date back more than 20 years, with one of the earliest studies describing the use of mRNA encoding the influenza virus nucleoprotein to generate a cytotoxic T cell response in mice5. Facilitated by potent mRNA delivery to antigen-presenting cells, there are ongoing clinical trials for mRNA vaccines for influenza, Zika, HIV and rabies.
Cancer immunotherapy. As with vaccines, mRNA-based cancer therapy requires the transfection of antigen-presenting cells. Although intradermal and intramuscular routes deliver mRNA to enough antigen-presenting cells to induce prophylactic immunity, their delivery efficiency is not high enough for some cancer immunotherapy applications. Instead, ex vivo transfection is used to activate high numbers of T cells capable of tackling well-established populations of tumour cells. There are two major types of mRNA cancer immunotherapy. The first functions at the cellular level in the same way as an mRNA vaccine except that the mRNA encodes tumour-associated peptide antigens. Numerous studies have shown that, following the transfection of dendritic cells with an mRNA cancer vaccine, cytotoxic T cells can target and destroy tumours in mice13,15,29,30.
The second class of mRNA-directed cancer immunotherapy, which involves the modification of T cells with chimeric antigen receptors (CARs), is called CAR T cell therapy31,32 (Fig. 4). Typically, a patient's T cells are isolated and transfected ex vivo with mRNA encoding CARs, which are protein fragments that are displayed on the T cell surface and bind to specific tumour epitopes33. Following the re-introduction of the modified T cells into patients, CARs home in on and kill tumour cells. For example, CARs specific for CD19, a B cell surface receptor, enable the cytotoxic T cell-mediated destruction of leukaemia in a xenograft mouse model34,35.
Cytotoxic T cells are transfected with mRNA encoding the chimeric antigen receptor (CAR) genes (step 1), which are translated into tumour antigen-specific receptor peptides (step 2). The expression of CARs on the T cell surface (step 3) facilitates tumour-specific targeting and subsequent cell lysis (step 4)161.
Gene editing. A particularly exciting application of mRNA therapeutics is gene editing, which involves the precision ‘cutting’ and ‘pasting’ of genomic DNA and a potentially permanent cure for gene-based diseases. The discovery of the celebrated CRISPR–Cas9 (CRISPR-associated protein 9) system — a bacterial defence mechanism adapted to generate double-stranded DNA breaks in eukaryotic cells36 — has led to the meteoric rise of the gene-editing field. CRISPR-mediated gene editing requires only two components: Cas9, a nuclease responsible for the DNA cleavage, and a short guide RNA (gRNA) that directs Cas9 to cut the DNA at a precise location37.
Typically, the two CRISPR components are delivered to cells using a DNA plasmid containing the genes for both the Cas9 protein and the gRNA38,39. One of the biggest limitations of plasmid-mediated gene editing is the potential to cut the genome at undesired, off-target locations as a result of prolonged expression of Cas9 in the cell40. As such, the co-delivery of gRNA and mRNA encoding Cas9 is an attractive alternative, as Cas9 expression is transient (Fig. 5). Although Cas9 is the nuclease currently used for most gene-editing applications, alternative nucleases, such as Cpf1 (Ref. 41), can also be delivered into the cell via mRNA42.
The target cell to be edited is transfected with mRNA encoding the Cas9 (CRISPR-associated protein 9) nuclease, which is translated by the ribosomal complex (step 1). Provided that the guide RNA (gRNA) is also transfected into the target cell (step 2), the Cas9 protein complexes with the gRNA to form a ribonucleoprotein with affinity for a specific DNA sequence (step 3). The ribonucleoprotein complex then translocates to the nucleus (step 4) where it binds to a DNA sequence complementary to the gRNA, mediating a double-stranded DNA ‘cut’ (step 5). The double-stranded DNA break is then either repaired by an error-prone mechanism called non-homologous end joining that can lead to a gene knockout or is replaced by a newly inserted piece of DNA in the presence of a ‘repair template’, a process known as homology-directed repair (step 6)37.
One of the most advanced applications of CRISPR–Cas9-mediated gene editing is the generation of enhanced CAR T cells for cancer immunotherapy (described above). In fact, the first planned human clinical trial using CRISPR involves the generation of modified CAR T cells. Although CAR T cells are typically generated from a patient's own cells, some patients’ T cells are not of sufficiently high quality43. Remarkably, CRISPR–Cas9 can be used to generate CAR T cells that are accepted by most patients by knocking out genes associated with graft-versus-host disease and the immune response to donor material43,44. CRISPR–Cas9 technology has also been used to improve the antitumour potency of CAR T cells45,46.
Cellular reprogramming. Of interest to the tissue engineering and regenerative medicine communities, mRNA can also be used to redirect cell fate and function. In a 2012 Nobel Prize winning discovery, Yamanaka and Gurdon showed that human somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) via the introduction of four transcription factors (octamer-binding transcription factor 4 (OCT4), SOX2, MYC and Krueppel-like factor 4 (KLF4))47. Cocktails of mRNA encoding these proteins, known as the Yamanaka factors, have been used to create iPSCs that lack the virus-mediated gene integration seen with plasmid delivery48,49. Similar strategies have reprogrammed other types of cell. For example, human vascular endothelial factor A (VEGFA)-encoding mRNA has been used to treat myocardial infarction10. Following intramyocardial injection of mRNA in mice, heart progenitor cells increasingly differentiated into endothelial cells, which resulted in improved heart function and long-term survival.
Design of in vitro-transcribed mRNA
Structural components. Native mRNA is a long, single-stranded polynucleotide with an average length of 2,000–2,500 bases50. It consists of five unique domains, each of which is important to include when designing IVT mRNA to ensure stability and translation (Fig. 6). The 5′ cap, otherwise known as the 7-methylguanosine cap (m7G), is a single methylated nucleoside affixed to the 5′ end of the mRNA51. As it is attached via a 5′ to 5′ triphosphate linkage, the 5′ end of the mRNA takes on the chemical appearance of what is normally the 3′ end of a nucleotide. The unique structure of the 5′ cap sterically protects the mRNA from degradation by exonucleases through association with cap-binding proteins52. Furthermore, the cap promotes translation initiation through its association with the eukaryotic initiation factor 4F (eIF4F) complex, which recruits ribosomes53.
The key structural domains of mRNA are the 5′ cap (also known as the 7-methylguanosine cap), the 5′ and 3′ untranslated regions (UTRs), the coding sequence and the poly(A) tail.
At the opposite end of mRNA, the 3′ poly(A) tail has a crucial role in regulating mRNA stability. As its name implies, the 3′ poly(A) tail is a chain of adenosine nucleotides on the 3′ end of the mRNA, with an estimated median length of 50–100 nucleotides in HeLa and NIH 3T3 cells54. Furthermore, the same study suggests that poly(A) tail length correlates with mRNA half-life but not translation efficiency54. For IVT mRNA, tail lengths of at least 30–40 adenosines have been deemed necessary to inhibit both 5′ to 3′ and 3′ to 5′ mRNA degradation pathways55,56, with 120 adenosines being optimal57.
Flanking the coding sequence, the 5′ and 3′ untranslated regions (UTRs) of mRNAs further affect translation and degradation. The nucleotides within these regions are not translated into protein; rather, specific sequences regulate translation of the coding sequence by binding to the eIF4F complex (at the 5′ end)58 and microRNAs (at the 3′ end). In addition, the 3′ UTR can contain AU-rich regions that promote mRNA decay59, which offers a mechanism by which to tune the duration of an mRNA drug's translation. As all but two amino acids (methionine and tryptophan) are encoded by multiple codons, mRNA sequence choice is somewhat flexible. When choosing UTR sequences for IVT mRNA, a common strategy involves the integration of UTR domains from human mRNAs, such as human β-globin, that naturally show high levels of translation and stability57,60.
Chemical base modifications. Although the chemical alteration of RNA bases may not seem like an obvious design requirement for IVT mRNA, it is necessary for most applications because it disguises artificially introduced mRNA from the immune system. Typically, human mRNA is produced in the nucleus and exported into the cytoplasm without interfacing with any other parts of the cell. The delivery of therapeutic mRNA, however, involves transport across the target cell membrane, often by an uptake process known as endocytosis61.
Interestingly, both immune and non-immune cells have specialized receptors that alert the immune system when RNA is detected outside of normal ‘bounds’, as foreign RNA is a hallmark of viral infection. In non-immune cells, a receptor called retinoic acid-inducible gene I protein (RIG-I) patrols the cell cytoplasm and, on recognition of RNA, can induce innate immune responses62,63. Furthermore, immune cells such as macrophages and dendritic cells have Toll-like receptors (TLRs) that, on binding to RNA, invoke a signalling cascade that causes inflammation, inhibited translation and/or RNA degradation64,65. Specifically, TLR3 detects RNA on the cell surface, and TLR7 and TLR8 ‘stake out’ the endosomal membrane66. GU-rich sequences are particularly susceptible to TLR recognition65.
Fortunately, it has been shown that the chemical modification of select IVT mRNA nucleotides reduces interactions with innate immune receptors, slowing clearance and substantially enhancing efficacy67. Various modifications have been assessed for reductions in TLR activation, including pseudouridine (ψ), N1-methylpseudouridine (m1ψ), 5-methoxyuridine (5moU), 5-methyluridine (m5U), 2-thiouridine (s2U), 5-methylcytidine (m5C), N1-methyladenosine (m1A) and N6-methyladenosine (m6A) (Fig. 7). Typically, modified mRNA sequences are synthesized by replacing all nucleotides of a particular native base with the corresponding modified base (for example, all uridines are replaced by pseudouridines). Importantly, base modification does not prohibit mRNA translation by ribosomes, as base modification is a naturally occurring post-transcriptional process in mammalian cells. It has been estimated that the m6A base, which is the most prevalent modified base in eukaryotes, occurs in the place of one in three adenosine residues in mammalian mRNA68.
Replacement of native nucleosides in in vitro-transcribed mRNA with a chemically modified version (or versions) reduces immunogenicity and increases translation efficiency.
On delivery to dendritic cells, m6A and s2U modifications have been shown to reduce TLR3 activation, and m5C, m5U, s2U, m6A and ψ limit TLR7 and TLR8 activation67,69. Of these, m5C and ψ are the most commonly used, with their incorporation facilitating up to a ninefold increase in translation in cell culture70. This was confirmed in vivo, with lower immunogenicity and higher stability observed following injection of modified mRNA in mice, particularly those incorporating pseudouridine71–73. A more recent study identified m1ψ as a potent base modification in vivo, with up to a 13-fold increase in gene expression observed compared with ψ or m5C/ψ (Ref. 74). Other strategies for reducing immunogenicity include high-performance liquid chromatography (HPLC) purification of IVT mRNA to remove any double-stranded RNA contaminants, which has been shown to increase translation in primary cells up to 1,000-fold compared with unpurified mRNA and to prevent induction of inflammatory cytokines75.
The need for delivery vehicles
Physical and chemical properties of mRNA. At 105–106 Da in size, mRNA is three to four orders of magnitude larger than molecules that readily diffuse into the cell, and its dense negative charge electrostatically repulses the anionic cell membrane. It has been shown that the cellular uptake rate of naked mRNA (that is, mRNA without a delivery vehicle) is less than 1 in 10,000 molecules4. With a median intracellular half-life of only 7 hours76, mRNA is also an inherently unstable molecule that is susceptible to degradation by 5′ exonucleases, 3′ exonucleases and endonucleases77. Together, these physical properties necessitate the use of a delivery vehicle both in vitro and in vivo.
In vitro transfection methods. Several utilitarian techniques have been developed to deliver mRNA and other nucleic acids into the cell cytoplasm for in vitro and ex vivo applications. Electroporation, one of the oldest means of transfection, increases cell membrane permeability to nucleic acids using an electrical field78. Electroporation can deliver naked mRNA for applications including protein replacement79, cancer immunotherapy80 and gene editing81. Another transfection method is the ‘gene gun’, which shoots metal (for example, gold) particles that have been surface coated with mRNA into the cell, propelled by pressurized carrier gas82. Gene guns were used for some of the earliest mRNA therapeutic studies, including vaccine development83,84. Microinjection, another physical mRNA transfection method, employs a micropipette to inject the nucleic acid directly into the intracellular space85.
So far, in the majority of clinical trials, mRNA is administered ex vivo using one of the above-mentioned techniques. Specifically, blood cells are removed from the patient, transfected in a cell culture dish and then re-infused. This approach ensures that sufficiently high numbers of target cells are transfected, which maximizes efficacy, and that only target cells are transfected, which minimizes side effects. Unfortunately, ex vivo therapy is costly and invasive compared with systemic injection86 and is usually limited to targeting white blood cells. Systemic delivery strategies are needed if the broad clinical potential of mRNA therapeutics is to be realized.
Systemic delivery barriers. Although localized injections have been used to deliver naked mRNA for prophylactic vaccines and cancer immunotherapies87–89, most therapeutic applications require systemic delivery. This can be accomplished by nanoparticulate vehicles that encapsulate mRNA and traverse a series of extracellular and intracellular barriers en route to the cytoplasm of the target cells. Following injection into the bloodstream, nanoparticles must evade immune cells and renal clearance by glomerular filtration while avoiding nonspecific interactions with serum proteins90,91. The vehicle must then cross the endothelial barrier and diffuse to the vicinity of the target tissue, which is challenging given that molecules larger than 5 nm typically do not cross the capillary endothelium (with the exception of some tissues such as the liver and spleen)92. Once in the extracellular matrix, the vehicle must diffuse through a network of fibrous proteins and polysaccharides to the target cell membrane93. After mediating cellular uptake, usually by endocytosis, a delivery vehicle must escape the endosome before unloading its mRNA cargo into the cytosol where translation can occur61. This process of endosomal escape may, indeed, be the most daunting aspect of delivery94, as it has been estimated that even world-class RNA delivery materials escape the endosome only ∼2% of the time95.
Materials for mRNA delivery
The identification of effective, nontoxic delivery systems remains one of the biggest challenges preventing the widespread clinical application of mRNA therapeutics. Although viral systems such as lentiviruses, adeno-associated viruses96 and the Sendai virus97 are capable of systemic nucleic acid delivery, their use can be limited by unwanted immune responses98. As such, this Review focuses on non-viral materials that have recently been efficacious in preclinical animal models (Table 1). Of note, many of the delivery materials discussed below have benefited from nearly two decades of research into non-viral short-interfering RNA (siRNA) delivery materials90,99. Although siRNA is double stranded, more rigid and much smaller than mRNA (∼42 bases compared with more than 1,000 bases), it is made of the same chemical building blocks and requires delivery to the same part of the cell: the cytoplasm.
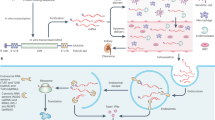
Lipids and lipid-like materials. A mainstay of non-viral nucleic acid delivery, liposomes, are spherical vesicles consisting of one (unilamellar) or more (multilamellar) phospholipid bilayers surrounding an aqueous core that houses the drug of interest (Fig. 8). They are typically prepared using materials that contain polar head groups and nonpolar tails, with the hydrophobic and hydrophilic interactions between these groups spurring vesicle formation100. The earliest liposomal delivery systems used cationic lipids such as DOTMA (1,2-di-O-octadecenyl-3-trimethylammonium-propane) that readily form complexes with anionic nucleic acids, including mRNA100,101. Other lipids such as the cationic DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) and zwitterionic DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) (Fig. 8b) have also been used alone or in combination with other materials for mRNA delivery102–104. The high degree of efficacy across diverse cell lines and the reproducibility of formulation that occurs with cationic liposomes have led to a host of commercial products (for example, lipofectamine) that have been used for mRNA delivery in vitro72,103 and in vivo10,72,73,105.
a | Lipid nanoparticles (LNPs) typically consist of an ionizable lipid, a helper lipid, cholesterol and polyethylene glycol (PEG) lipid. The mRNA is incorporated into the hydrophilic interior of the nanoparticle. b | Lipid and lipid-like materials developed for in vivo mRNA delivery7,121,127. Also included is the structure of DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), a helper lipid that imparts efficacy to mRNA–LNP formulations119. c | Polymeric nanomicelles are typically formulated from a block co-polymer in which the hydrophobic domain forms an outer shell and the hydrophilic domain forms an inner core that electrostatically complexes with anionic mRNA. d | Polymeric materials that have been developed for in vivo mRNA delivery134,138. Also included are the structures of linear and branched polyethylenimine (PEI), which have been modified for potent mRNA delivery11,132,133.
Although attractive from an efficacy perspective, cationic lipid-based delivery systems are associated with toxicity and immunogenicity in vitro and in vivo. For example, intravenously injected cationic liposomes can cause liver damage106 and can elicit a strong interferon-γ response in mice, resulting in inflammation107,108. Furthermore, positively charged lipids such as DOTAP and DOTMA can be neutralized by anionic serum proteins, leading to toxicity and reduced efficacy109.
As an alternative, ionizable lipidic systems have been developed that reduce the toxicity induced by cationic particles while retaining their advantageous transfection characteristics. Specifically, effective ionizable lipids tend to be positively charged at low pH (which aids in RNA complexation when it is carried out in acidic buffer) but are neutral at physiological pH (for reduced toxicity post injection). Furthermore, cellular uptake via endocytosis deposits nanoparticles into endosomal compartments, which slowly reduce their pH from approximately 6.8 to 4.5 as they morph into lysosomes110. An ability for nanoparticles to ionize as the pH drops seems to be crucial to the endosomal escape process. Although mechanisms are not yet well understood, it is thought that the lipid's positive charge facilitates its electrostatic interaction and subsequent fusion with the negatively charged endosomal membrane111. This fusion destabilizes the lipid bilayer, resulting in discharge of the nucleic acid cargo into the cytoplasm112,113. The importance of the ability of nucleic acid delivery systems to take on positive charge under mildly acidic conditions is underscored by studies that report efficacious siRNA delivery only for lipid nanoparticle chemistries with surface pKa values ranging from 5.5 to 7 (Refs 114,115).
In addition to a charged or ionizable lipid, lipid nanoparticle (LNP) formulations typically include cholesterol, a helper lipid and a polyethylene glycol (PEG) lipid (Fig. 8a). Cholesterol, which is hydrophobic and rigid, fills in the gaps between lipids within liposomal membranes, promoting vesicle stability116. Helper lipids such as 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and DOPE enhance LNP efficacy by promoting fusion with both cell and endosomal membranes, facilitating cell uptake and endosomal release117. Finally, PEG lipids consist of a PEG molecule conjugated to alkyl chains that anchor themselves into the LNP bilayer. Of particular importance for systemic delivery, PEG lipids reduce opsonization by serum proteins as well as reticuloendothelial clearance118.
The precise formulation of the LNP — that is, the ratio of ionizable lipid to cholesterol to helper lipid to PEG lipid — can substantially affect LNP efficacy. Compared with LNP formulations designed for siRNA delivery, optimal mRNA delivery has been shown to require lower amounts of ionizable lipid and cholesterol and higher amounts of helper lipid and PEG lipid119. Replacing the helper lipid DSPC, a common compound in siRNA LNP formulations, with DOPE also confers high levels of mRNA delivery efficacy. This may be because unsaturated lipids, such as DOPE, can form unstable hexagonal rather than stable lamellar phases, leading to reduced membrane stability and enhanced endosomal escape119,120.
Most LNP-mediated mRNA delivery studies have focused on protein replacement applications that include experimentally convenient end points. For example, the ionizable lipid-like material OF-02 (Fig. 8b) was shown to systemically deliver erythropoietin mRNA, which is one of the few therapeutic IVT mRNAs that are commercially available121. Erythropoietin, a protein produced by the kidneys and readily detected in the blood, is relevant to the treatment of anaemia.
LNPs similar in composition to those previously reported by Alnylam Pharmaceuticals114 have been used to examine the influence of route of administration on mRNA delivery122. LNPs loaded with modified mRNA encoding firefly luciferase were administered by intravenous, subcutaneous, intradermal, intramuscular and intraperitoneal injection as well as by intratracheal instillation. Intravenous administration resulted in the highest luminescence signal (predominantly in the liver) but the shortest duration of expression. Intramuscular and intradermal injections offered the best duration of effect, with protein production maintained locally 10 days post injection. Similar LNPs facilitated HIV protection by mRNA delivery. A single intravenous injection of 1.4 mg kg−1 of mRNA encoding the light and heavy chains of the VRC01 HIV-1-specific antibody protected two different forms of humanized mice when challenged with HIV only 1 or 2 weeks after mRNA delivery12.
Protein replacement studies have attempted treatment of haemophilia B with mRNA encoding factor IX. Factor IX, which is a blood-clotting factor normally produced in hepatocytes, is easily quantified from blood serum samples. In one study, LNPs formulated with the lipid-like material C12-200 were used to deliver mRNA to factor IX-knockout mice. A single dose of LNPs containing 0.5 mg kg−1 factor IX mRNA, administered intravenously, yielded therapeutically relevant protein levels 12 hours after injection8. A similar effect was seen with a lipid-like material based on N,N,N-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide (TT3), which contains a central phenyl ring, three amide linkers and three amino lipid chains7 (Fig. 8b). Systemic delivery of optimized LNPs complexed with 1.1 mg kg−1 human factor IX mRNA restored normal protein levels. A proprietary LNP formulation developed by Arcturus Therapeutics has also been shown to potently deliver factor IX mRNA9. Tail vein injections of 2 mg kg−1 mRNA loaded into LNPs induced therapeutic levels of factor IX for 4–6 days after administration.
Although many preclinical studies involving LNPs have focused on protein replacement, other applications have also been reported. In the area of cancer immunotherapy, the subcutaneous injection of LNPs carrying mRNA encoding the B16F10 melanoma tumour antigens glycoprotein 100 (gp100) and tyrosine-related protein 2 (TRP2) resulted in transfection of antigen-presenting cells in vivo. Reductions in tumour volume and extended survival were observed as a result of CD8+ T cell activation13.
mRNA vaccines are being developed using proprietary LNP technology, with two recent studies describing the use of LNPs for Zika vaccination. In a collaboration involving the University of Pennsylvania and Acuitas Therapeutics, LNPs were formulated with mRNA encoding the Zika pre-membrane and envelope (prM-E)123. A single intradermal immunization of LNPs encapsulating 30 μg of prM-E mRNA protected mice from Zika virus challenges at 2 weeks and 5 months post injection. Non-human primates survived a challenge 5 weeks after immunization with as little as 50 μg of prM-E mRNA. A second study from the University of Washington and Valera (a Moderna Therapeutics subsidiary) described protection against Zika viral challenge as a result of high neutralizing antibody titres following intramuscular injection of two doses of prM-E mRNA LNPs in mice. In this study, mice were immunized with 10 μg of prM-E mRNA loaded into LNPs, with a further booster dose given at 3 weeks, followed by Zika virus challenge 6 weeks after the initial dose124. Moderna has also reported on an mRNA avian influenza vaccine encoding haemagglutinin proteins, which are glycoproteins found on the surface of influenza viruses. Mice were protected from a lethal dose of virus following intradermal immunization with as little as 0.4 μg of mRNA and non-human primates developed a robust immune response when immunized intradermally or intramuscularly125.
LNPs can also deliver CRISPR–Cas9 RNAs for in vivo gene editing. For example, a hybrid viral–non-viral system facilitated gene editing in mouse hepatocytes that corrected a mutation responsible for human hereditary tyrosinaemia. C12-200 LNPs carrying Cas9 mRNA were systemically co-delivered with a viral vector carrying a gRNA and a repair template, leading to repair of the mutated gene and alleviation of disease symptoms126. One of the first studies to achieve gene editing in vivo using only non-viral delivery materials used zwitterionic amino lipids127. LNPs formulated from these modified lipids, which contain a sulfobetaine head group, amine-rich linkers and hydrophobic tails, co-delivered Cas9 mRNA and gRNA in vitro and in vivo. The most effective material, ZA3-EP10 (Fig. 8b), facilitated 95% luciferase gene knockout in cell culture, as well as potent gene editing in mouse liver, kidneys and lungs following systemic delivery of 5 mg kg−1 total RNA. Notably, efficacy was shown both when mRNA and gRNA delivery was ‘staged’ (with gRNA delivery occurring hours after Cas9 mRNA delivery) and when they were co-delivered. Co-delivery is more challenging because the gRNA must remain stable and available within the cytoplasm of the cell until the Cas9 mRNA is translated into functional protein hours later. The lipid-like material TT3 (Fig. 8b) is another non-viral material capable of delivering all of the CRISPR components needed to edit genes128. Staged, systemic delivery of Cas9 mRNA-loaded TT3 LNPs (0.56 mg kg−1) followed 6 hours later by gRNA-loaded LNPs (0.25 mg kg−1) mediated the deletion of the hypercholesterolemia-related gene proprotein convertase subtilisin–kexin-type 9 (Pcsk9) in mouse livers.
Polymers. Although not as clinically advanced as lipid systems for mRNA delivery, polymers have shown considerable potential in protein replacement, vaccine and other applications related to mRNA therapeutics. Cationic polymers shuttle nucleic acids across cellular and subcellular membranes by condensing them into nanoplexes. Both linear and branched cationic polymers can facilitate cellular uptake by endocytosis and subsequent endosomal escape129. Polyethylenimine (PEI) (Fig. 8d) is one of the best-established polymers for nucleic acid delivery130,131. However, its broad therapeutic application has been thwarted by toxicity associated with high molecular weight (>25 kDa) and highly branched formulations109. Similarly to cationic lipids, this toxicity has been attributed to the interaction of cationic polymers with negatively charged serum proteins, such as albumin, which cause the nanoplexes to aggregate, increasing their effective size.
To mitigate some of these issues, some researchers have used low molecular weight modified PEI for mRNA delivery. For example, a polymer synthesized from stearic acid and branched 2 kDa PEI was used to deliver mRNA encoding HIV-1 gag. Following subcutaneous injection, immunoglobulin G (IgG) antibody titres were higher in mice that had received the treatment compared with a delivery vehicle control11. In another study, 2 kDa PEI conjugated to cyclodextrin was used for the intranasal administration of HIV antigen mRNA, which resulted in a strong systemic and mucosal HIV-specific immune response132. Recently, the primary amines of branched 1.8 kDa PEI with aromatic domains were modified for improved mRNA delivery in HeLa and U87 cells compared with native 1.8 kDa PEI133.
An alternative to PEI, self-assembled polymeric nanomicelles (Fig. 8c) are made of block copolymers in which a polyamino acid block forms an inner core surrounded by an outer PEG ‘shell’ with mRNA complexed electrostatically in the core. For example, the polymer poly(N′-(N-(2-aminoethyl)-2-aminoethyl)aspartamide), known as PAsp(DET), is commonly combined with PEG to form the block copolymer PEG-PAsp(DET) (Fig. 8d). PEG-PAsp(DET) can transfect nasal neurons with mRNA following intranasal administration for protein replacement therapy134. Furthermore, hydrodynamic injection with PEG-PAsp(DET) nanomicelles complexed with mRNA encoding the anti-apoptotic protein B cell lymphoma-2 (BCL-2) mitigated apoptosis in a mouse model of fulminant hepatitis135.
Other variations on PEG-PAsp block copolymers, including PEG-PAsp(TET) and PEG-PAsp(TEP), have been used to deliver mRNA. Local injection of both PEG-PAsp(DET) and PEG-PAsp(TET) nanomicelles loaded with transcription factor mRNA into mouse knee joints slowed disease progression in a mouse osteoarthritis model136. In another study, PEG-PAsp(TEP)–cholesterol particles were formulated with mRNA encoding the anti-angiogenic protein FMS-like tyrosine kinase 1 (FLT1; also known as VEGFR1). Their systemic injection (once every 2 days for 14 days) significantly inhibited tumour growth in a subcutaneous mouse model of pancreatic cancer14. PEG-poly(lysine) block copolymers have also been used for tumour-targeted mRNA-mediated protein replacement therapy. For example, PEG-poly(lysine) particles decorated with tumour-specific ligands resulted in significant protein expression in tumours after systemic delivery of mRNA in mice137.
In 2016, Dong and co-workers reported on the synthesis and testing of a polymer brush library consisting of alkyl tails conjugated to poly(glycoamidoamines) backbones. When formulated with cholesterol, DSPC and PEG lipid, the material TarN3C10 (Fig. 8d), which contained a tartarate sugar backbone and 10-carbon long tails, most potently delivered mRNA and siRNA in vivo138. In a separate study, systemic delivery of PEGylated poly(β-amino esters) induced luciferase expression in mouse lungs139. The data show disparities that can arise between nanoparticle biodistribution and the location of protein production, underscoring the importance of cell uptake and mRNA translation capacity across organs139.
Polymeric nanoparticles have also been used to generate antigen-specific immunity for vaccine applications. A dendrimer-based nanoparticle system has been developed to deliver antigen-encoding replicon mRNA, which is a type of self-amplifying mRNA. Mice immunized intramuscularly with a single dose of mRNA-loaded dendrimer nanoparticles mounted cellular and antibody responses that protected them from lethal challenges with H1N1 influenza and Ebola viruses as well as with the Toxoplasma gondii parasite140.
Charge-altering releasable transporters (CARTs), which are polymers that contain a cationic oligo(α-amino ester) domain and a lipophilic block, have been used for systemic mRNA delivery. It is postulated that CARTs, which are cationic at low pH, undergo a physical rearrangement upon cellular entry that neutralizes the cationic charge, leading to decomplexation and release of the mRNA into the cytoplasm. Systemic delivery of the most effective CART carrying luciferase mRNA mediated potent protein expression in the liver and spleen, peaking 4 hours post injection141. In another recent report, various oligoalkylamines grafted to poly(acrylic acid) were used to generate a small library of polymeric nanoparticles for luciferase mRNA delivery. The lead polymer, PAA20k-(2-3-2), induced significant protein expression in pig lung tissue upon pulmonary delivery142.
Hybrid systems. Transcript-activated matrices (TAMs) are a unique mRNA delivery system made of a biomaterial matrix loaded with mRNA lipoplexes that undergo controlled release. TAMs incorporating mRNA encoding bone morphogenetic protein 2 (BMP2), a protein involved in bone and cartilage formation, in a collagen sponge improved femur bone regeneration relative to unloaded sponges in rats143. In a more recent study, TAMs were made by loading mRNA lipoplexes on either fibrin gel or biphasic calcium phosphate granules for cellular reprogramming. Stem cells transfected with either type of TAM promoted osteogenic differentiation and the calcium phosphate granule system induced more mineral deposition144.
Hybrid graphene oxide–PEI complexes have been used for mRNA-based cellular reprogramming. Graphene oxide–PEI complexed with mRNA encoding the Yamanaka transcription factors facilitated the generation of iPSCs from tissue-derived fibroblasts145. In addition, an mRNA delivery system has been developed that uses eIF4E, a protein that initiates translation. To enhance translation and decrease mRNA degradation on cellular entry, mRNA was complexed with eIF4E along with a cationic polyamine carrier to facilitate delivery. The resulting nanoplexes transfected dendritic cells ex vivo for immunotherapy and induced luciferase expression in mouse lungs on systemic administration146.
Other considerations
Kinetics of mRNA translation. The rate at which proteins are synthesized from IVT mRNA is an important consideration when assessing mRNA delivery systems. Although most studies report peak protein translation within 6 hours of administration121,122,139,141, others analyse 24 hour time points127,142. In a study of mRNA translation kinetics, fluorescence microscopy tracked individual A549 cells following their transfection with GFP-encoding mRNA using the transfection reagent Lipofectamine 2000 (Ref. 147). GFP production was observed as early as 5 hours, with peak expression occurring 20 hours post transfection.
The mRNA coding sequence also affects the translation kinetics and efficiency. Alteration of a single nucleotide in the coding sequence without changing the resultant amino acid sequence was found to affect protein structure because of distorted folding during translation148. Furthermore, it was found that integration of rare codons into the coding sequence reduced the rate of translation because these rare codons require the recruitment of unique tRNAs149. More extensive studies are needed to describe how the kinetics of translation vary with delivery vehicle, target cell type and encoded protein.
Acquiring IVT mRNA. The procurement of reasonably priced mRNA appropriate for use in delivery studies has been a considerable obstacle. In vitro transcription kits, which have become increasingly commercially available, enable individual laboratories to synthesize their own mRNA at increased yields and reduced cost. When purchasing a kit, it is crucial that the promoter sequence on the vector to be transcribed matches the polymerase of choice. Most kits use enzymes compatible with the T7 promoter9,75,122,141 but others include the SP6 and T3 polymerases150. Kits have also been developed to include capping and polyadenylating enzymes for the one-pot synthesis of fully capped and polyadenylated mRNA14,57. Alternatively, capping and polyadenylation reactions can be carried out post-transcriptionally using specialized kits. Modified mRNAs with reduced immunogenicity can also be in vitro-transcribed by replacing unmodified nucleotides with the modified version (for example, replacing uridine-5′-triphosphate with pseudouridine-5′-triphosphate) in the IVT reaction at the desired ratio. Finally, transcript purification must be accomplished carefully to rid IVT mRNA products of contaminants, such as double-stranded RNA species, that promote immune recognition and decrease efficacy in vivo75.
Currently, the primary commercial supplier of presynthesized mRNA is TriLink Biotechnologies. Although it offers modified and unmodified mRNAs encoding several reporter, gene editing and therapeutic proteins, selection is limited for most therapeutic applications. Furthermore, the cost of purchasing commercially synthesized mRNA is high, with modified mRNAs costing on the order of US$1 per μg (as a point of reference, some siRNAs are an order of magnitude less expensive). As such, well-controlled in vivo experiments can be prohibitively expensive using commercially available options. An mRNA dose of 1 mg kg−1 to an average-sized mouse, for example, would cost approximately $20. Although the cost of using IVT kits is generally lower on a per microgram basis, considerable man-hours are required to produce high-quality mRNAs in house.
Clinical translation of mRNA therapeutics
Most mRNA clinical trials over the past 20 years have relied on the ex vivo transfection and re-infusion of immune cells, mainly for cancer immunotherapy applications. More than 20 clinical trials involving this strategy are currently active (Table 2). Although most of these trials involve the classic strategy of transfusing autologous dendritic cells electroporated with tumour-associated antigens, several phase I/II trials carried out in China are using sequencing technology to identify patient-specific tumour antigens for personalized brain cancer immunotherapies. In other trials, T cells are transfected ex vivo with tumour-targeting CARs for the treatment of B cell leukaemia, B cell lymphoma and mesothelioma. Another phase I trial being conducted at the University of Pennsylvania involves the transfusion of T cells that have been genetically engineered ex vivo for HIV vaccines. In this trial, autologous CD4+ T cells are transfected with zinc finger nuclease mRNA, resulting in editing of CC-chemokine receptor 5 (CCR5), a receptor that HIV uses to enter host cells. Re-administration of these cells back into patients has the potential to generate a population of HIV-immune T cells.
Several clinical studies are attempting in vivo delivery of mRNA therapeutics. For example, Moderna Therapeutics, has four ongoing or planned clinical trials using proprietary LNP technology for vaccines. Each vaccine, delivered intramuscularly, contains mRNA encoding either influenza, chikungunya or Zika virus antigen. Initial data from their H10 influenza trial indicate robust prophylactic immunity in humans after antigen mRNA immunization125. Other clinical studies for vaccine development include a phase I trial by CurVac AG for the rabies virus, as well as trials at the Fundació Clínic per la Recerca Biomèdica, Barcelona, Spain, and the Erasmus Medical Center, Rotterdam, Netherlands, for HIV vaccines. Moderna also has plans to carry out two trials for cancer immunotherapy, one in collaboration with Merck. Finally, AstraZeneca, in collaboration with Moderna, has filed a clinical trial application to initiate a phase I trial using VEGFA-encoding mRNA formulations to promote angiogenesis in cardiac patients151.
Conclusions and future directions
mRNA holds significant promise as a genetic medicine, avoiding several key issues associated with traditional DNA-based gene therapy. Perhaps most importantly, mRNA induces only transient protein expression, which can facilitate a wide range of biological processes without the risk of genomic integration. Applications such as immunotherapy, gene editing and cell reprogramming require protein expression for only limited periods of time. The two overarching factors that have hindered the clinical progression of mRNA therapeutics are the instability and immunogenicity of IVT mRNA and the lack of sufficiently efficacious delivery systems. The challenge of instability and immunogenicity of IVT mRNA is expected to be addressed as we learn more about how the design of the untranslated regions and poly(A) tail and the inclusion of modified bases affect the stability and translation efficiency of IVT mRNA. On the delivery front, the most clinically advanced materials take the form of lipid nanoparticles. Unfortunately, because the majority of these delivery formulations are proprietary, more academic studies are needed to help the broader scientific community understand how lipid structure and nanoparticle formulation affect efficacy and influence potential applications. That being said, there is cause for optimism: with the right delivery materials, mRNA therapeutics have the potential to revolutionize medicine as we know it, facilitating personalized, gene-based therapies that are as unique as the patients for whom they are intended.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
How to cite this article
Hajj, K. A. & Whitehead, K. A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2, 17056 (2017).
References
Vila, A., Sanchez, A., Tobio, M. & Calvo, P. Design of biodegradable particles for protein delivery. J. Control. Release 78, 15–24 (2002).
Friedmann, T. & Roblin, R. Gene therapy for human genetic disease? Science 175, 949–955 (1972).
Cavazzana, M., Six, E., Lagresle-Peyrou, C., André-Schmutz, I. & Hacein-Bey-Abina, S. Gene therapy for X-linked severe combined immunodeficiency: where do we stand? Hum. Gene Ther. 27, 108–116 (2016).
Sahin, U., Kariko, K. & Türeci, Ö . mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014).
Martinon, F. et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 23, 1719–1722 (1993). This paper is one of the earliest studies using liposome-entrapped mRNA for an influenza vaccine.
Conry, R., LoBuglio, A., Wright, M. & Sumerel, L. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 55, 1397–1400 (1995).
Li, B. et al. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 15, 8099–8107 (2015).
DeRosa, F. et al. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 23, 699–707 (2016).
Ramaswamy, S. et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl Acad. Sci. USA 114, E1941–E1950 (2017).
Zangi, L. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 31, 898–907 (2013).
Zhao, M., Li, M., Zhang, Z., Gong, T. & Sun, X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 23, 2596–2607 (2016).
Pardi, N. et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 8, 14630 (2017). Describes protection against HIV-1 challenge in mice using lipid nanoparticles carrying mRNA encoding HIV-specific antibodies.
Oberli, M. A. et al. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 17, 1326–1335 (2016).
Uchida, S. et al. Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials 82, 221–228 (2016).
Broos, K. et al. Particle-mediated intravenous delivery of antigen mRNA results in strong antigen-specific T cell responses despite the induction of type I interferon. Mol. Ther. Nucleic Acids 5, e326 (2016).
Josephson, N. The hemophilias and their clinical management. Hematology Am. Soc. Hematol. Educ. Program 2013, 261–267 (2013).
Bonne, G. et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat. Genet. 21, 285–288 (1999).
Crick, F. On protein synthesis. Symp. Soc. Exp. Biol. 12, 138–163 (1958).
Warner, J., Knopf, P. & Rich, A. A multiple ribosomal structure in protein synthesis. Proc. Natl Acad. Sci. USA 49, 122–129 (1963).
Wolff, J. A. et al. Direct gene transfer into mouse muscle in vivo. Science 247, 1465–1468 (1990). Shows that naked mRNA injected into mice is translated.
Jirikowski, G., Sanna, P. P., Maciejewski-Lenoir, D. & Bloom, F. Reversal of diabetes insipidus in Brattleboro rats: intrathypothalmic injection of vasopressin mRNA. Science 255, 996–998 (1992). Shows alleviation of disease symptoms using mRNA-mediated protein replacement.
Wong, C.-H. Protein glycosylation: new challenges and opportunities. J. Org. Chem. 70, 4219–4225 (2005).
Khoury, G. A., Baliban, R. C. & Floudas, C. A. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 90 (2011).
Schlake, T., Thess, A., Fotin-Mleczek, M. & Kallen, K.-J. Developing mRNA-vaccine technologies. RNA Biol. 9, 1319–1330 (2012).
Midoux, P. & Pichon, C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccines 14, 221–234 (2015).
Deering, R. P., Kommareddy, S., Ulmer, J. B., Brito, L. A. & Geall, A. J. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 11, 885–899 (2014).
Malissen, B., Tamoutounour, S. & Henri, S. The origins and functions of dendritic cells and macrophages in the skin. Nat. Rev. Immunol. 14, 417–428 (2014).
Langlet, C. et al. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J. Immunol. 188, 1751–1760 (2012).
Nair, S., Heiser, A., Boczkowski, D. & Majumdar, A. Induction of cytotoxic T cell responses and tumour immunity against unrelated tumours using telomerase reverse transcriptase RNA transfected dendritic cells. Nat. Med. 6, 1011–1017 (2000).
Perche, F. et al. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumour antigen messenger RNA. Nanomedicine 7, 445–453 (2011).
Eshhar, Z., Waks, T., Bendavid, A. & Schindler, D. Functional expression of chimeric receptor genes in human T cells. J. Immunol. Methods 248, 67–76 (2001).
Kalos, M. & June, C. H. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity 39, 49–60 (2013).
Hegde, M. et al. Combinational targeting offsets antigen escape and enhances effector functions of adoptively transferred T Cells in glioblastoma. Mol. Ther. 21, 2087–2101 (2013).
Barrett, D. M. et al. Treatment of advanced leukemia in mice with mRNA engineered T cells. Hum. Gene Ther. 22, 1575–1586 (2011).
Barrett, D. M. et al. Regimen-specific effects of RNA-modified chimeric antigen receptor T cells in mice with advanced leukemia. Hum. Gene Ther. 24, 717–727 (2013).
Wiedenheft, B., Sternberg, S. H. & Doudna, J. A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 (2012).
Doudna, J. A. & Charpentier, E. The new frontier of genome engineering with CRISPR–Cas9. Science 346, 1258096 (2014).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR –Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell 163, 759–771 (2015).
Kim, Y. et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 34, 808–810 (2016).
Liu, X. et al. CRISPR–Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 27, 154–157 (2016).
Ren, J. et al. Multiplex genome editing to generate universal CAR T Cells resistant to PD1 inhibition. Clin. Cancer Res. 23, 2255–2266 (2017).
Rupp, L. J. et al. CRISPR–Cas9-mediated PD-1 disruption enhances anti-tumour efficacy of human chimeric antigen receptor T cells. Sci. Rep. 7, 737 (2017).
Eyquem, J. et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Warren, L. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010). Reports the generation of iPSC from somatic cells using mRNA.
Mandal, P. K. & Rossi, D. J. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc. 8, 568–582 (2013).
Ota, T. et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36, 40–45 (2004).
Shatkin, A. J. Capping of eucaryotic mRNAs. Cell 9, 645–653 (1976).
Evdokimova, V., Ruzanov, P., Imataka, H., Raught, B. & Svitkin, Y. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20, 5491–5502 (2001).
Haghighat, A. & Sonenberg, N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272, 21677–21680 (1997).
Chang, H., Lim, J., Ha, M. & Kim, V. N. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53, 1044–1052 (2014).
Guhaniyogi, J. & Brewer, G. Regulation of mRNA stability in mammalian cells. Gene 265, 11–23 (2001).
Meijer, H. A. et al. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 35, e132 (2007).
Holtkamp, S. et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108, 4009–4017 (2006).
Mignone, F., Gissi, C., Liuni, S. & Pesole, G. Untranslated regions of mRNAs. Genome Biol. 3, reviews0004-1 (2002).
Chen, C.-Y. & Shyu, A.-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470 (1995).
Kariko, K., Kuo, A. & Barnathan, E. S. Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA. Gene Ther. 6, 1092–1100 (1999).
Lorenz, C. et al. Protein expression from exogenous mRNA: uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol. 8, 627–636 (2011).
Hornung, V., Ellegast, J., Kim, S., Brzozka, K. & Jung, A. 5′-Triphosphate RNA Is the ligand for RIG-I. Science 314, 994–997 (2006).
Loo, Y.-M. & Gale, M. Jr. Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 (2011).
Diebold, S., Kaisho, T., Hemmi, H., Akira, S. & Sousa, C. R. E. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004).
Heil, F., Hemmi, H., Hochrein, H., Ampenberger, F. & Kirschning, C. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303, 1526–1529 (2004).
Lee, B. L. & Barton, G. M. Trafficking of endosomal Toll-like receptors. Trends Cell Biol. 24, 360–369 (2014).
Kariko, K. & Weissman, D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implications for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 10, 523–532 (2007).
Yao, B. et al. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17, 537–549 (2016).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). Reports that nucleoside-modified mRNA is non-immunogenic.
Kariko, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008).
Anderson, B. R. et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010).
Kormann, M. S. D. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 (2011).
Kariko, K., Muramatsu, H., Keller, J. M. & Weissman, D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 20, 948–953 (2012).
Andries, O. et al. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344 (2015).
Kariko, K., Muramatsu, H., Ludwig, J. & Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 39, e142 (2011).
Sharova, L. V. et al. Database for mRNA half-life of 19,977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 16, 45–58 (2008).
Houseley, J. & Tollervey, D. The many pathways of RNA degradation. Cell 136, 763–776 (2009).
Neumann, E., Schaefer-Ridder, M., Wang, Y. & Hofschneider, P. H. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1, 841–845 (1982).
Smits, E. et al. RNA-based gene transfer for adult stem cells and T cells. Leukemia 18, 1898–1902 (2004).
Van Tendeloo, V. F. I. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumour antigen loading of dendritic cells. Blood 98, 49–56 (2001).
Hashimoto, M. & Takemoto, T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep. 5, 11315 (2015).
Klein, T. M., Wolf, E. D., Wu, R. & Sanford, J. C. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 327, 70–73 (1987).
Qiu, P., Ziegelhoffer, P., Sun, J. & Yang, N. S. Gene gun delivery of mRNA in situ results in efficient transgene and genetic immunization. Gene Ther. 3, 262–268 (1996).
Mandl, C. W. et al. In vitro-synthesized infectious RNA as an attenuated live vaccine in a flavivirus model. Nat. Med. 4, 1438–1440 (1998).
Murata-Hori, M. & Wang, Y.-L. Microinjection of mRNA into somatic cells. eLShttp://dx.doi.org/10.1038/npg.els.0002696 (2005).
Wang, T., Upponi, J. R. & Torchilin, V. P. Design of multifunctional non-viral gene vectors to overcome physiological barriers: dilemmas and strategies. Int. J. Pharm. 427, 3–20 (2012).
Roesler, E. et al. Immunize and disappear — safety-optimized mRNA vaccination with a panel of 29 allergens. J. Allergy Clin. Immunol. 124, 1070–1077 (2009).
Petsch, B. et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol 30, 1210–1216 (2012).
Hattinger, E. et al. Prophylactic mRNA vaccination against allergy confers long-term memory responses and persistent protection in mice. J. Immunol. Res. 2015, 797421 (2015).
Whitehead, K. A., Langer, R. & Anderson, D. G. Knocking down barriers; advances in siRNA delivery. Nat. Rev. Drug Discov. 8, 129–138 (2009).
Yin, H. et al. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014).
Alexis, F., Pridgen, E., Molnar, L. K. & Farokhzad, O. C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 5, 505–515 (2008).
Zamecnik, J., Vargova, L., Homola, A., Kodet, R. & Sykova, E. Extracellular matrix glycoproteins and diffusion barriers in human astrocytic tumours. Neuropathol. Appl. Neurobiol. 30, 338–350 (2004).
Wittrup, A. et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 33, 870–876 (2015).
Gilleron, J. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638–646 (2013).
Giacca, M. & Zacchigna, S. Virus-mediated gene delivery for human gene therapy. J. Control. Release 161, 377–388 (2012).
Ramani, K., Hassan, Q., Venkaiah, B., Hasnain, S. & Sarkar, D. Site-specific gene delivery in vivo through engineered Sendai viral envelopes. Proc. Natl Acad. Sci. USA 95, 11886–11890 (1998).
Nayak, S. & Herzog, R. W. Progress and prospects: immune responses to viral vectors. Gene Ther. 17, 295–304 (2010).
Tatiparti, K., Sau, S., Kashaw, S. & Iyer, A. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials 7, 77 (2017).
Torchilin, V. P. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 4, 145–160 (2005).
Malone, R. W., Felgner, P. L. & Verma, I. M. Cationic liposome-mediated RNA transfection. Proc. Natl Acad. Sci. USA 86, 6077–6081 (1989).
Anderson, D. M. et al. Stability of mRNA/cationic lipid lipoplexes in human and rat cerebrospinal fluid: methods and evidence for nonviral mRNA gene delivery to the central nervous system. Hum. Gene Ther. 14, 191–202 (2003).
Rejman, J., Tavernier, G., Bavarsad, N., Demeester, J. & De Smedt, S. C. mRNA transfection of cervical carcinoma and mesenchymal stem cells mediated by cationic carriers. J. Control. Release 147, 385–391 (2010).
Kranz, L. M. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016).
Thess, A. et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 23, 1456–1464 (2015).
Landesman-Milo, D. & Peer, D. Toxicity profiling of several common RNAi-based nanomedicines: a comparative study. Drug Deliv. Transl Res. 4, 96–103 (2014).
Ma, Z. et al. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem. Biophys. Res. Commun. 330, 755–759 (2005).
Platanias, L. C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005).
Lv, H., Zhang, S., Wang, B., Cui, S. & Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 114, 100–109 (2006).
Huotari, J. & Helenius, A. Endosome maturation. EMBO J. 30, 3481–3500 (2011).
Walsh, C. L., Nguyen, J., Tiffany, M. R. & Szoka, F. C. Synthesis, characterization, and evaluation of ionizable lysine-based Lipids for siRNA delivery. Bioconjug. Chem. 24, 36–43 (2013).
Zelphati, O. & Szoka, F. Mechanism of oligonucleotide release from cationic liposomes. Proc. Natl Acad. Sci. USA 93, 11493–11498 (1996).
Hafez, I. M., Maurer, N. & Cullis, P. R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 8, 1188–1196 (2001).
Jayaraman, M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. 51, 8529–8533 (2012).
Whitehead, K. A. et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun 5, 4277 (2014).
Cheng, X. & Lee, R. J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 99, 129–137 (2016).
Li, W. & Szoka, F. C. Jr. Lipid-based nanoparticles for nucleic acid delivery. Pharm. Res. 24, 438–449 (2007).
Jokerst, J. V., Lobovkina, T., Zare, R. N. & Gambhir, S. S. Nanoparticle PEGylation for imaging and therapy. Nanomedicine 6, 715–728 (2011).
Kauffman, K. J. et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 15, 7300–7306 (2015).
Harvie, P., Wong, F. & Bally, M. Characterization of lipid DNA interactions. I. Destabilization of bound lipids and DNA dissociation. Biophys. J. 75, 1040–1051 (1998).
Fenton, O. S. et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater 28, 2939–2943 (2016).
Pardi, N. et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release 217, 345–351 (2015).
Pardi, N. et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 543, 248–251 (2017). Shows Zika virus protection in animals using lipid nanoparticle–mRNA formulation.
Richner, J. M. et al. Modified mRNA vaccines protect against Zika virus infection. Cell 168, 1114–1125 (2017).
Bahl, K. et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 25, 1316–1327 (2017).
Yin, H. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol 34, 328–333 (2016).
Miller, J. B. et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. 56, 1059–1063 (2017). Reports the non-viral, systemic delivery of CRISPR RNAs for gene editing in mice.
Jiang, C. et al. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 27, 440–443 (2017).
Akinc, A., Thomas, M., Klibanov, A. M. & Langer, R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 7, 657–663 (2004).
Boussif, O., Lezoualch, F., Zanta, M. A. & Mergny, M. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA 92, 7297–7301 (1995).
Lungwitz, U., Breunig, M., Blunk, T. & Göpferich, A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 60, 247–266 (2005).
Li, M. et al. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Control. Release 228, 9–19 (2016).
Chiper, M., Tounsi, N., Kole, R., Kichler, A. & Zuber, G. Self-aggregating 1.8 kDa polyethylenimines with dissolution switch at endosomal acidic pH are delivery carriers for plasmid DNA, mRNA, siRNA and exon-skipping oligonucleotides. J. Control. Release 246, 60–70 (2017).
Baba, M., Itaka, K., Kondo, K., Yamasoba, T. & Kataoka, K. Treatment of neurological disorders by introducing mRNA in vivo using polyplex nanomicelles. J. Control. Release 201, 41–48 (2015).
Matsui, A., Uchida, S., Ishii, T., Itaka, K. & Kataoka, K. Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases. Sci. Rep. 5, 15810 (2015).
Aini, H. et al. Messenger RNA delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci. Rep. 6, 18743 (2016).
Chen, Q. et al. A targeted and stable polymeric nanoformulation enhances systemic delivery of mRNA to tumours. Mol. Ther. 25, 92–101 (2017).
Dong, Y. et al. Poly(glycoamidoamine) brushes formulated nanomaterials for systemic siRNA and mRNA delivery in vivo. Nano Lett. 16, 842–848 (2016).
Kaczmarek, J. C. et al. Polymer–lipid nanoparticles for systemic delivery of mRNA to the lungs. Angew. Chem. Int. Ed. 128, 14012–14016 (2016).
Chahal, J. S. et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl Acad. Sci. USA 113, E4133–E4142 (2016).
McKinlay, C. J. et al. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc. Natl Acad. Sci. USA 114, E448–E456 (2017).
Jarzçbin´ska, A. et al. A single methylene group in oligoalkylamine-based cationic polymers and lipids promotes enhanced mRNA delivery. Angew. Chem. Int. Ed. 55, 9591–9595 (2016).
Badieyan, Z. S. et al. Transcript-activated collagen matrix as sustained mRNA delivery system for bone regeneration. J. Control. Release 239, 137–148 (2016).
Balmayor, E. R. et al. Modified mRNA for BMP-2 in combination with biomaterials serves as a transcript-activated matrix for effectively inducing osteogenic pathways in stem cells. Stem Cells Dev. 26, 25–34 (2017).
Choi, H. Y. et al. Efficient mRNA delivery with graphene oxide–polyethylenimine for generation of footprint-free human induced pluripotent stem cells. J. Control. Release 235, 222–235 (2016).
Li, J. et al. Structurally programmed assembly of translation initiation nanoplex for superior mRNA delivery. ACS Nano 11, 2531–2544 (2017).
Leonhardt, C. et al. Single-cell mRNA transfection studies: delivery, kinetics and statistics by numbers. Nanomedicine 10, 679–688 (2014).
Kimchi-Sarfaty, C. et al. A ‘silent’ polymorphism in MDR1 gene changes substrate specificity. Science 315, 525–528 (2007).
Cannarozzi, G. et al. A role for codon order in translation dynamics. Cell 141, 355–367 (2010).
Pokrovskaya, I. & Gurevich, V. In vitro transcription: preparative RNA yields in analytical scale reactions. Anal. Biochem. 220, 420–423 (1994).
Moderna Therapeutics. Moderna Provides Pipeline and Full-Year Corporate Update. Moderna [online], https://www.modernatx.com/newsroom/press-releases/moderna-provides-pipeline-and-full-year-corporate-update (2017).
Brenner, S., Jacob, F. & Meselson, M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 180, 576–581 (1961). This is the first study to report on the discovery of mRNA.
Lockard, R. E. & Lingrel, J. B. The synthesis of mouse hemoglobin beta-chains in a rabbit reticulocyte cell-free system programmed with mouse reticulocyte 9S RNA. Biochem. Biophys. Res. Commun. 37, 204–212 (1969).
Dimitriadis, G. Translation of rabbit globin mRNA introduced by liposomes into mouse lymphocytes. Nature 274, 923–924 (1978). This is the first report of mRNA delivery using liposomes.
Zhou, W. Z. et al. RNA melanoma vaccine: induction of antitumour immunity by human glycoprotein 100 mRNA immunization. Hum. Gene Ther. 10, 2719–2724 (1999).
Heiser, A. et al. Autologous dendritic cells transfected with prostate-specific antigen RNA stimulate CTL responses against metastatic prostate tumours. J. Clin. Invest. 109, 409–417 (2002).
Doyon, Y. et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 26, 702–708 (2008).
Yoon, S. H. et al. Adoptive immunotherapy using human peripheral blood lymphocytes transferred with RNA encoding Her-2/neu-specific chimeric immune receptor in ovarian cancer xenograft model. Cancer Gene Ther. 16, 489–497 (2009).
Wood, A. J. et al. Targeted genome editing across species using ZFNs and TALENs. Science 333, 307–307 (2011).
Hwang, W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013).
Maus, M., Grupp, S. A., Porter, D. L. & June, C. H. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood 123, 2625–2635 (2014).
Luo, X. et al. Dual-functional lipid-like nanoparticles for delivery of mRNA and MRI contrast agents. Nanoscale 9, 1575–1579 (2017).
Mahiny, A. J. et al. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat. Biotechnol. 33, 584–586 (2015).
Lin, C.-Y. et al. Messenger RNA-based therapeutics for brain diseases: an animal study for augmenting clearance of β-amyloid by intracerebral administration of neprilysin mRNA loaded in polyplex nanomicelles. J. Control. Release 235, 268–275 (2016).
Acknowledgements
Funding was provided by the Defense Advanced Research Projects Agency (DARPA), grant number D16AP00143. The authors thank R. Weiss for his feedback on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Hajj, K., Whitehead, K. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat Rev Mater 2, 17056 (2017). https://doi.org/10.1038/natrevmats.2017.56
Published:
DOI: https://doi.org/10.1038/natrevmats.2017.56
This article is cited by
-
CD19/CD20 dual-targeted chimeric antigen receptor-engineered natural killer cells exhibit improved cytotoxicity against acute lymphoblastic leukemia
Journal of Translational Medicine (2024)
-
Combinatorial development of nebulized mRNA delivery formulations for the lungs
Nature Nanotechnology (2024)
-
mRNA-based vaccines and therapeutics: an in-depth survey of current and upcoming clinical applications
Journal of Biomedical Science (2023)
-
Emerging non-viral vectors for gene delivery
Journal of Nanobiotechnology (2023)
-
Lipid nanoparticle-based mRNA delivery systems for cancer immunotherapy
Nano Convergence (2023)