Abstract
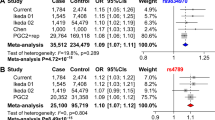
Bipolar disorder (BPD), schizophrenia (SCZ) and unipolar major depressive disorder (MDD) are primary psychiatric disorders sharing substantial genetic risk factors. We previously reported that two single-nucleotide polymorphisms (SNPs) rs2709370 and rs6785 in the cAMP responsive element-binding (CREB)-1 gene (CREB1) were associated with the risk of BPD and abnormal hippocampal function in populations of European ancestry. In the present study, we further expanded our analyses of rs2709370 and rs6785 in multiple BPD, SCZ and MDD data sets, including the published Psychiatric Genomics Consortium (PGC) genome-wide association study, the samples used in our previous CREB1 study, and six additional cohorts (three new BPD samples, two new SCZ samples and one new MDD sample). Although the associations of both CREB1 SNPs with each illness were not replicated in the new cohorts (BPD analysis in 871 cases and 1089 controls (rs2709370, P=0.0611; rs6785, P=0.0544); SCZ analysis in 1273 cases and 1072 controls (rs2709370, P=0.230; rs6785, P=0.661); and MDD analysis in 129 cases and 100 controls (rs2709370, P=0.114; rs6785, P=0.188)), an overall meta-analysis of all included samples suggested that both SNPs were significantly associated with increased risk of BPD (11 105 cases and 51 331 controls; rs2709370, P=2.33 × 10−4; rs6785, P=6.33 × 10−5), SCZ (34 913 cases and 44 528 controls; rs2709370, P=3.96 × 10−5; rs6785, P=2.44 × 10−5) and MDD (9369 cases and 9619 controls; rs2709370, P=0.0144; rs6785, P=0.0314), with the same direction of allelic effects across diagnostic categories. We then examined the impact of diagnostic status on CREB1 mRNA expression using data obtained from independent brain tissue samples, and observed that the mRNA expression of CREB1 was significantly downregulated in psychiatric patients compared with healthy controls. The protein–protein interaction analyses showed that the protein encoded by CREB1 directly interacted with several risk genes of psychiatric disorders identified by GWAS. In conclusion, the current study suggests that CREB1 might be a common risk gene for major psychiatric disorders, and further investigations are necessary.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Barch DM. Neuropsychological abnormalities in schizophrenia and major mood disorders: similarities and differences. Curr Psychiatry Rep 2009; 11: 313–319.
Benabarre A, Vieta E, Colom F, Martinez-Aran A, Reinares M, Gasto C. Bipolar disorder, schizoaffective disorder and schizophrenia: epidemiologic, clinical and prognostic differences. Eur Psychiatry 2001; 16: 167–172.
Greenwood TA. Positive traits in the bipolar spectrum: the space between madness and genius. Mol Neuropsychiatry 2017; 2: 198–212.
McGuffin P, Katz R. The genetics of depression and manic-depressive disorder. Br J Psychiatry 1989; 155: 294–304.
McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 2003; 60: 497–502.
Berrettini WH. Are schizophrenic and bipolar disorders related? A review of family and molecular studies. Biol Psychiatry 2000; 48: 531–538.
Grigoroiu-Serbanescu M, Rietschel M, Hauser J, Czerski PM, Herms S, Sun X et al. Commingling analysis of age-of-onset in bipolar I disorder and the morbid risk for major psychoses in first degree relatives of bipolar I probands. J Affect Disord 2014; 168: 197–204.
Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373: 234–239.
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379.
Craddock N, Owen MJ. The Kraepelinian dichotomy - going, going... but still not gone. Br J Psychiatry 2010; 196: 92–95.
Moskvina V, Craddock N, Holmans P, Nikolov I, Pahwa JS, Green E et al. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry 2009; 14: 252–260.
Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H et al. Molecular and genetic characterization of depression: overlap with other psychiatric disorders and aging. Mol Neuropsychiatry 2015; 1: 1–12.
Chang H, Hoshina N, Zhang C, Ma Y, Cao H, Wang Y et al. The protocadherin 17 gene affects cognition, personality, amygdala structure and function, synapse development and risk of major mood disorders. Mol Psychiatry 2017; DOI:10.1038/mp.2016.231.
Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, Webster MJ. Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol Psychiatry 2011; 69: 353–359.
Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry 2010; 15: 1016–1022.
Liu Y, Blackwood DH, Caesar S, de Geus EJ, Farmer A, Ferreira MA et al. Meta-analysis of genome-wide association data of bipolar disorder and major depressive disorder. Mol Psychiatry 2011; 16: 2–4.
McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet 2010; 42: 128–131.
Li L, Chang H, Peng T, Li M, Xiao X. Evidence of AS3MTd2d3-associated variants within 10q24.32-33 in the genetic risk of major affective disorders. Mol Neuropsychiatry 2017; 2: 213–218.
O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 2008; 40: 1053–1055.
Ou J, Li M, Xiao X. The schizophrenia susceptibility gene ZNF804A confers risk of major mood disorders. World J Biol Psychiatry 2017; 18: 557–562.
Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159.
Williams HJ, Craddock N, Russo G, Hamshere ML, Moskvina V, Dwyer S et al. Most genome-wide significant susceptibility loci for schizophrenia and bipolar disorder reported to date cross-traditional diagnostic boundaries. Hum Mol Genet 2011; 20: 387–391.
Chang H, Xiao X, Li M. The schizophrenia risk gene ZNF804A: clinical associations, biological mechanisms and neuronal functions. Mol Psychiatry 2017; 22: 944–953.
Liu Z, Huang L, Luo XJ, Wu L, Li M. MAOA variants and genetic susceptibility to major psychiatric disorders. Mol Neurobiol 2016; 53: 4319–4327.
Tang J, Fan Y, Li H, Xiang Q, Zhang D-F, Li Z et al. Whole-genome sequencing of monozygotic twins discordant for schizophrenia indicates multiple genetic risk factors for schizophrenia. J Genet Genomics 2017; 44: 295–306.
International Schizophrenia ConsortiumInternational Schizophrenia ConsortiumPurcell SM International Schizophrenia ConsortiumWray NR International Schizophrenia ConsortiumStone JL International Schizophrenia ConsortiumVisscher PM International Schizophrenia ConsortiumO'Donovan MC et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
Steinberg S, de Jong S Irish Schizophrenia Genomics ConsortiumAndreassen OA, Werge T, Borglum AD et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet 2011; 20: 4076–4081.
Li M, Wang Y, Zheng XB, Ikeda M, Iwata N, Luo XJ et al. Meta-analysis and brain imaging data support the involvement of VRK2 (rs2312147) in schizophrenia susceptibility. Schizophr Res 2012; 142: 200–205.
Chang H, Zhang C, Xiao X, Pu X, Liu Z, Wu L et al. Further evidence of VRK2 rs2312147 associated with schizophrenia. World J Biol Psychiatry 2016; 17: 457–466.
Chen X, Lee G, Maher BS, Fanous AH, Chen J, Zhao Z et al. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry 2011; 16: 1117–1129.
Zhang R, Zhang H, Li M, Li H, Li Y, Valenzuela RK et al. Genetic analysis of common variants in the CMYA5 (cardiomyopathy-associated 5) gene with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 2013; 46: 64–69.
Li M, Luo XJ, Zhang X, Yang ZH, Xiang K, Xiao X et al. A common variant of the cardiomyopathy associated 5 gene (CMYA5) is associated with schizophrenia in Chinese population. Schizophr Res 2011; 129: 217–219.
Wang Q, He K, Li Z, Chen J, Li W, Wen Z et al. The CMYA5 gene confers risk for both schizophrenia and major depressive disorder in the Han Chinese population. World J Biol Psychiatry 2014; 15: 553–560.
Hoshina N, Tanimura A, Yamasaki M, Inoue T, Fukabori R, Kuroda T et al. Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron 2013; 78: 839–854.
Li M, Luo XJ, Rietschel M, Lewis CM, Mattheisen M, Muller-Myhsok B et al. Allelic differences between Europeans and Chinese for CREB1 SNPs and their implications in gene expression regulation, hippocampal structure and function, and bipolar disorder susceptibility. Mol Psychiatry 2014; 19: 452–461.
Carlezon WA Jr., Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci 2005; 28: 436–445.
Weeber EJ, Levenson JM, Sweatt JD. Molecular genetics of human cognition. Mol Interv 2002; 2: 376–91, 39.
Wallace TL, Stellitano KE, Neve RL, Duman RS. Effects of cyclic adenosine monophosphate response element binding protein overexpression in the basolateral amygdala on behavioral models of depression and anxiety. Biol Psychiatry 2004; 56: 151–160.
Zubenko GS, Hughes HB 3rd, Maher BS, Stiffler JS, Zubenko WN, Marazita ML. Genetic linkage of region containing the CREB1 gene to depressive disorders in women from families with recurrent, early-onset, major depression. Am J Med Genet 2002; 114: 980–987.
Blendy JA. The role of CREB in depression and antidepressant treatment. Biol Psychiatry 2006; 59: 1144–1150.
Perlis RH, Purcell S, Fagerness J, Cusin C, Yamaki L, Fava M et al. Clinical and genetic dissection of anger expression and CREB1 polymorphisms in major depressive disorder. Biol Psychiatry 2007; 62: 536–540.
Psychiatric Gwas Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983.
Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun 2014; 5: 3339.
Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry 2009; 14: 755–763.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427.
Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009; 460: 753–757.
Major Depressive Disorder Working Group of the Psychiatric Gwas ConsortiumMajor Depressive Disorder Working Group of the Psychiatric Gwas ConsortiumRipke S Major Depressive Disorder Working Group of the Psychiatric Gwas ConsortiumWray NR Major Depressive Disorder Working Group of the Psychiatric Gwas ConsortiumLewis CM Major Depressive Disorder Working Group of the Psychiatric Gwas ConsortiumHamilton SP Major Depressive Disorder Working Group of the Psychiatric Gwas ConsortiumWeissman MM et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013; 18: 497–511.
Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011; 1: 457–470.
Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods 2011; 9: 179–181.
Genomes Project ConsortiumGenomes Project ConsortiumAbecasis GR Genomes Project ConsortiumAuton A Genomes Project ConsortiumBrooks LD Genomes Project ConsortiumDePristo MA Genomes Project ConsortiumDurbin RM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65.
Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 2007; 81: 1084–1097.
International HapMap ConsortiumInternational HapMap ConsortiumAltshuler DM International HapMap ConsortiumGibbs RA International HapMap ConsortiumPeltonen L International HapMap ConsortiumAltshuler DM International HapMap ConsortiumGibbs RA et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–58.
Li M, Chang H, Xiao X. BDNF Val66Met polymorphism and bipolar disorder in European populations: a risk association in case-control, family-based and GWAS studies. Neurosci Biobehav Rev 2016; 68: 218–233.
Xiao X, Luo XJ, Chang H, Liu Z, Li M. Evaluation of European Schizophrenia GWAS loci in Asian Populations via comprehensive meta-analyses. Mol Neurobiol 2016; 54: 4071–4080.
Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007; 3: 1724–1735.
Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011; 98: 152–153.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012; 28: 882–883.
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8: 118–127.
Luo X, Huang L, Jia P, Li M, Su B, Zhao Z et al. Protein-protein interaction and pathway analyses of top schizophrenia genes reveal schizophrenia susceptibility genes converge on common molecular networks and enrichment of nucleosome (chromatin) assembly genes in schizophrenia susceptibility loci. Schizophr Bull 2014; 40: 39–49.
Wu Y, Yao YG, Luo XJ. SZDB: a database for schizophrenia genetic research. Schizophr Bull 2017; 43: 459–471.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43(Database issue): D447–D452.
Dupont WD, Plummer WD Jr. Power and sample size calculations. A review and computer program. Control Clin Trials 1990; 11: 116–128.
Lanz TA, Joshi JJ, Reinhart V, Johnson K, Grantham LE 2nd, Volfson D. STEP levels are unchanged in pre-frontal cortex and associative striatum in post-mortem human brain samples from subjects with schizophrenia, bipolar disorder and major depressive disorder. PLoS ONE 2015; 10: e0121744.
Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol Psychiatry 2006; 11: 965–978.
Narayan S, Tang B, Head SR, Gilmartin TJ, Sutcliffe JG, Dean B et al. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res 2008; 1239: 235–248.
Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry 2004; 9: 406–416.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45: 580–585.
GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015; 348: 648–660.
Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014; 17: 1418–1428.
Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci 2012; 15: 1723–1728.
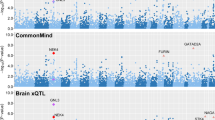
Network Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18: 199–209.
Mitchell AC, Javidfar B, Pothula V, Ibi D, Shen EY, Peter CJ et al. MEF2C transcription factor is associated with the genetic and epigenetic risk architecture of schizophrenia and improves cognition in mice. Mol Psychiatry 2017.
Blizinsky KD, Diaz-Castro B, Forrest MP, Schurmann B, Bach AP, Martin-de-Saavedra MD et al. Reversal of dendritic phenotypes in 16p11.2 microduplication mouse model neurons by pharmacological targeting of a network hub. Proc Natl Acad Sci USA 2016; 113: 8520–8525.
Harrington AJ, Raissi A, Rajkovich K, Berto S, Kumar J, Molinaro G et al. MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. Elife 2016 doi: 10.7554/eLife.20059.
Focking M, Lopez LM, English JA, Dicker P, Wolff A, Brindley E et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol Psychiatry 2015; 20: 424–432.
Erk S, Mohnke S, Ripke S, Lett TA, Veer IM, Wackerhagen C et al. Functional neuroimaging effects of recently discovered genetic risk loci for schizophrenia and polygenic risk profile in five RDoC subdomains. Transl Psychiatry 2017; 7: e997.
Kalkman HO. Potential opposite roles of the extracellular signal-regulated kinase (ERK) pathway in autism spectrum and bipolar disorders. Neurosci Biobehav Rev 2012; 36: 2206–2213.
Melas PA, Lennartsson A, Vakifahmetoglu-Norberg H, Wei Y, Aberg E, Werme M et al. Allele-specific programming of Npy and epigenetic effects of physical activity in a genetic model of depression. Transl Psychiatry 2013; 3: e255.
Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron 2002; 35: 605–623.
Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G et al. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron 1998; 21: 869–883.
Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J 2007; 26: 2284–2294.
Dyson HJ, Wright PE. Role of intrinsic protein disorder in the function and interactions of the transcriptional coactivators CREB-binding protein (CBP) and p300. J Biol Chem 2016; 291: 6714–6722.
Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 2000; 95: 745–755.
Kumar G, Clark SL, McClay JL, Shabalin AA, Adkins DE, Xie L et al. Refinement of schizophrenia GWAS loci using methylome-wide association data. Hum Genet 2015; 134: 77–87.
Juhasz G, Dunham JS, McKie S, Thomas E, Downey D, Chase D et al. The CREB1-BDNF-NTRK2 pathway in depression: multiple gene-cognition-environment interactions. Biol Psychiatry 2011; 69: 762–771.
Ma L, Wu DD, Ma SL, Tan L, Chen X, Tang NL et al. Molecular evolution in the CREB1 signal pathway and a rare haplotype in CREB1 with genetic predisposition to schizophrenia. J Psychiatr Res 2014; 57: 84–89.
Sokolowski M, Wasserman J, Wasserman D. Polygenic associations of neurodevelopmental genes in suicide attempt. Mol Psychiatry 2016; 21: 1381–1390.
Barrot M, Olivier JD, Perrotti LI, DiLeone RJ, Berton O, Eisch AJ et al. CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proc Natl Acad Sci USA 2002; 99: 11435–11440.
Yamada S, Yamamoto M, Ozawa H, Riederer P, Saito T. Reduced phosphorylation of cyclic AMP-responsive element binding protein in the postmortem orbitofrontal cortex of patients with major depressive disorder. J Neural Transm (Vienna) 2003; 110: 671–680.
Nibuya M, Nestler EJ, Duman RS. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J Neurosci 1996; 16: 2365–2372.
Jeon SH, Seong YS, Juhnn YS, Kang UG, Ha KS, Kim YS et al. Electroconvulsive shock increases the phosphorylation of cyclic AMP response element binding protein at Ser-133 in rat hippocampus but not in cerebellum. Neuropharmacology 1997; 36: 411–414.
Chen AC, Shirayama Y, Shin KH, Neve RL, Duman RS. Expression of the cAMP response element binding protein (CREB) in hippocampus produces an antidepressant effect. Biol Psychiatry 2001; 49: 753–762.
O'Donnell JM, Zhang HT. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4). Trends Pharmacol Sci 2004; 25: 158–163.
Fleischhacker WW, Hinterhuber H, Bauer H, Pflug B, Berner P, Simhandl C et al. A multicenter double-blind study of three different doses of the new cAMP-phosphodiesterase inhibitor rolipram in patients with major depressive disorder. Neuropsychobiology 1992; 26: 59–64.
Pliakas AM, Carlson RR, Neve RL, Konradi C, Nestler EJ, Carlezon WA Jr. Altered responsiveness to cocaine and increased immobility in the forced swim test associated with elevated cAMP response element-binding protein expression in nucleus accumbens. J Neurosci 2001; 21: 7397–7403.
Barco A, Pittenger C, CREB KandelER. memory enhancement and the treatment of memory disorders: promises, pitfalls and prospects. Expert Opin Ther Targets 2003; 7: 101–114.
Acknowledgments
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No., XDB13000000); Chinese Academy of Sciences (CAS) Pioneer Hundred Talents Program (to ML); National Natural Science Foundation of China (81471358); and Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (20152530). The Romanian sample recruitment and genotyping was funded by UEFISCDI, Bucharest, Romania (grant no. 89/2012 to MG-S) and by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (grant 01ZX1314A). The data of USA samples used for the analyses described in this manuscript were obtained from dbGaP accession number phs000979.v1.p1; the data of GAIN AA sample used for the analyses described in this manuscript were obtained from dbGaP accession number phs000021.v3.p2 (schizophrenia) and phs000017.v3.p1 (bipolar disorder). This research was supported by the Intramural Research Program of the NIMH (NCT00001260, 900142).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Xiao, X., Zhang, C., Grigoroiu-Serbanescu, M. et al. The cAMP responsive element-binding (CREB)-1 gene increases risk of major psychiatric disorders. Mol Psychiatry 23, 1957–1967 (2018). https://doi.org/10.1038/mp.2017.243
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2017.243
This article is cited by
-
Correlation between variants of the CREB1 and GRM7 genes and risk of depression
BMC Psychiatry (2023)
-
CaMKK2 as an emerging treatment target for bipolar disorder
Molecular Psychiatry (2023)
-
Identification of schizophrenia symptom-related gene modules by postmortem brain transcriptome analysis
Translational Psychiatry (2023)
-
Inactivation of ERK1/2-CREB Pathway Is Implicated in MK801-induced Cognitive Impairment
Current Medical Science (2023)
-
Polymorphisms of COMT and CREB1 are associated with treatment-resistant depression in a Chinese Han population
Journal of Neural Transmission (2022)