Abstract
Tourette Disorder (TD) is a childhood-onset neuropsychiatric and neurodevelopmental disorder characterized by the presence of both motor and vocal tics. The genetic architecture of TD is believed to be complex and heterogeneous. Nevertheless, DNA sequence variants co-segregating with TD phenotypes within multiplex families have been identified. This report examines whole exomes of affected and unaffected individuals in a multiplex TD family to discover genes involved in the TD etiology. We performed whole exome sequencing on six out of nine members in a three-generation TD multiplex family. Putative deleterious sequence variants co-segregating with TD patients were identified by our in-house bioinformatics pipeline. Induced pluripotent stem cells (iPSCs) were generated from one unaffected and two TD affected individuals. Neurons were derived from the iPSCs and biochemical assays were conducted to evaluate possible molecular differences between affected and unaffected. A rare heterozygous nonsense mutation in PNKD was co-segregated with TD in this multiplex family. Transcript and protein levels of the PNKD long isoform were reduced in neurons derived from the individuals with TD due to the nonsense mutation, indicating nonsense-mediated mRNA decay. We demonstrated that the PNKD long isoform monomer oligomerizes with itself as well as interacts with the synaptic active zone protein RIMS1α. We concluded that reduced PNKD long isoform levels are detected in all affected individuals and we provide evidence for a mechanism whereby this might contribute to the TD phenotype.
Similar content being viewed by others
Introduction
Tourette Disorder (TD) is a heritable early-onset neurodevelopmental disorder characterized by the presence of both motor and vocal tics with unclear neuropathogenesis as well as complex genetic architecture.1 The etiology of TD appears to be polygenic and heterogeneous, probably involving dozens of common variants with small effect size or rare variants with larger effect size.2 An initial genome-wide association study did not identify common variants meeting significance thresholds.3 However, a linkage study on a multiplex family identified a rare dominant-negative nonsense mutation (W317X) at the L-histidine decarboxylase (HDC) gene.4 In addition, targeted sequencing of the region where chromosomal abnormalities were found in a TD patient identified the SLITRK1 gene, in which rare recurrent variants were further identified.5 To identify potential TD-associated rare sequence variant(s) with large effect size, we focused on whole exome sequencing (WES) of multiplex families. In one particular family heavily affected by TD and Tic disorders, a novel nonsense mutation within the paroxysmal nonkinesigenic dyskinesia (PNKD) gene was identified, which appeared to be a strong TD candidate.
To evaluate the impact(s) that the PNKD nonsense mutation might have on gene expression and cellular functions, we generated induced pluripotent stem cells (iPSCs) and differentiated them into neuronal cells in vitro. With the iPSC-derived neurons from unaffected and affected family members, we were able to measure the endogenous transcript level and protein expression of the neuronal-specific PNKD long (L) isoform and we examined its subcellular localization. In addition, we demonstrated the self-oligomerization of the PNKD (L) protein monomer and its interaction with synaptic protein RIMS1α in transfected human cells and their co-localization in the human iPSC-derived neurons.
Materials and methods
Human subjects
A three-generation multiplex family was recruited through the Tourette International Collaborative Genetics (TIC Genetics)6 study and New Jersey Center for Tourette Syndrome (NJCTS)7 with informed consent by all participants. The study protocol has been approved by Rutgers University Institutional Review Board.
The whole exome sequencing and sanger sequencing
Genomic DNA was extracted from the bloods of subjects 3001, 3002, 4001, 4002, 5001, 5003 and 5005 and the saliva of subject 5004. whole exome sequencing was performed on subjects 4001, 4002, 5001, 5003 (proband), 5004 and 5005 (Figure 1a). For the WES library preparation, the SureSelect Human All Exon V5+UTR kit (Agilent Technologies, Santa Clara, CA, USA) was used to capture the targeted region for sequencing on the Illumina HiSeq2000. Variants were called by Geospiza’s GeneSifter (PerkinElmer, Waltham, MA, USA). For Sanger sequencing, the genomic region harboring the nonsense mutation was PCR-amplified with primers shown in Supplementary Table S1.
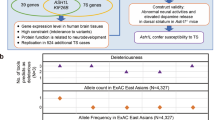
A rare heterozygous nonsense mutation (C to T transition) was identified in a TD multiplex family by whole exome sequencing. (a) We performed whole exome sequencing on 4001, 4002, 5001, 5003 (proband), 5004 and 5005. (b) The heterozygous nonsense mutation at the PNKD gene was present in subjects 3001, 4001, 5001, 5003, 5004 and 5005. TD, Tourette disorder.
pVAAST analysis
The pedigree Variant Annotation, Analysis and Search Tool (pVAAST) program prioritized candidate genes8 under both dominant and recessive modes of inheritance (see supplement).
Generation and maintenance of iPSCs
Induced pluripotent stem cells were generated from the CD4+ T lymphocytes using CytoTune-iPS reprogramming kit.9 Induced pluripotent stem cells were cultured on Matrigel-coated plates in mTeSR (see supplement).
Generation and maintenance of neural progenitor cells and neurons
neural progenitor cells were generated from iPSCs using a neural rosette-based protocol. Neurons were derived from NPCs by culturing the NPCs in neural differentiation medium (see supplement).
Real-time quantitative PCR
Total RNA was extracted using the RNeasy Mini kit (QIAGEN, Hilden, Germany). cDNA was made from 1ug total RNA using TaqMan Reverse Transcription Reagents (Thermo Fisher Scientific, Waltham, MA, USA). Real-time quantitative PCR (RT-qPCR) was performed on ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Primer information is shown in Supplementary Table S1.
Plasmids construction and transfection
Human PNKD (L) cDNA was cloned from cDNA ORF Clone (RC206179) (Origene, Rockville, MD, USA) and was inserted into the pcDNA4/TO vector. By PCR, a FLAG tag or a Myc tag was added to the PNKD (L) cDNA sequence as shown: 5′-FLAG-PNKD-3′ or 5′- PNKD-Myc-3′. To express human RIMS1α, RIMS1 transcript variant 1, cDNA ORF Clone (RC213013) (Origene) was used. The expression of GFP cloned into the pcDNA4/TO vector monitored transfection efficiency. To transfect neurons plated on poly-L-ornithine/laminin/fibronectin-coated 12 mm coverslips, we used Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). To transfect 293FT (Thermo Fisher Scientific) cells plated in 10 cm dishes, we used 1 mg ml−1 PEI solution (Sigma-Aldrich, St. Louis, MO, USA). A room temperature mixture of plasmids, transfection reagents and Opti-MEM reduced-serum medium were added to the cultures. Two days later, cells were collected for RNA extraction, or lysed in protein lysis buffer or fixed for immunocytochemistry.
Western blot and co-immunoprecipitation
Cells were lysed in RIPA buffer for western blot analysis or in RIPA IP buffer for co-immunoprecipitation. Anti-FLAG M2 Magnetic beads (Sigma-Aldrich) eluted FLAG-tagged proteins. Total protein lysates or eluates of the anti-FLAG M2 beads were resolved on NuPAGE 10% Bis-Tris or 3–8% Tris-acetate gels (Thermo Fisher Scientific).
Electrophysiology
Whole cell path clamp experiments were performed as described elsewhere.10, 11 Whole cell patch clamp recordings were performed in human neurons derived from iPSCs in oxygenated artificial cerebrospinal fluid at 30 °C. The artificial cerebrospinal fluid contains (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 2.5 Glucose, 22.5 Sucrose, 2.5 CaCl2, 1.2 MgCl2. The whole-cell pipette solution contained (in mM) 126 K-Gluconate, 4 KCl, 10 HEPES, 0.3 Na-GTP, 4 Mg-adenosine triphosphate (ATP), 0.05 ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid (EGTA) and 10 phosphocreatine (pH 7.2, adjusted with KOH). When whole cell currents were recorded, voltage clamp mode was used, and cells were held at −70 mV and a depolarization protocol from −100 to +90 mV in 10 mV intervals were applied. When spontaneous potentials were recorded, current clamp I=0 was used. Whole cell patch clamp recordings were performed using an Axon 700B amplifier. Data were filtered at 2 kHz, digitized at 10 kHz and collected using Clampex 10.2 (Molecular Devices, Sunnyvale, CA, USA).
Results
Clinical evaluations of the TD multiplex family
In this nonconsanguineous three-generation pedigree (Figure 1a), the proband (5003) and all four of her full siblings participated, as well as both parents and paternal grandparents (see supplement for clinical data).
A rare nonsense mutation in PNKD is associated with TD or Tic disorder
We performed WES on individual 4001, 4002, 5001, 5003 (proband), 5004 and 5005, generating more than 60 million 100 bp paired-end reads for each, with an average coverage of 53 × (Supplementary Table S3). We identified about 200 000 variants per individual. From these variants, we prioritized candidate genes using the pVAAST program, which identifies genes that contain highly differentiated functional variants between the cases and a reference control population of normal individuals. pVAAST also directly incorporates the variant inheritance pattern within the family into the likelihood ratio test for gene prioritization.8 A total of 145 and 169 genes were scored under dominant and recessive modes of inheritance, respectively (Supplementary Table S4). Under the dominant mode, PNKD has the highest score. Tourette or Tic Disorder-affected family members shared a heterozygous nonsense mutation (chr2: 219204814 C/T), which is not present in the unaffected individuals and absent in the background sample. By Sanger sequencing, we confirmed the heterozygous C to T transition in PNKD is also present in subject 3001, and validated the genotypes in 4001, 5001, 5003, 5004 and 5005 (an example is shown in Figure 1b). The Exome Aggregation Consortium (ExAC) database12 indicates that this variant was found in 4 of 120 802 alleles, giving an allele frequency of 3.3 × 10−5. In our recent study where WES of 511 TD trios were completed,13 this PNKD nonsense mutation was not identified, suggesting its rarity.
Missense mutations in PNKD are associated with familial PNKD, a neurological movement disorder causing episodic involuntary movement attacks.14, 15, 16 Because of the deleterious nature of the nonsense mutation, its segregation pattern within the pedigree, and the neurological movement disorder associated with PNKD, we selected PNKD for further investigation.
Neuronal cells were generated from the human subjects through iPSC intermediates
We generated NPCs and subsequently neurons from iPSCs of three members (4002-Uaf; 5001-TD1; 5003 (proband)-TD2) and one unrelated unaffected subject (Urel_Uaf) without the mutation (Supplementary Figure S6A). Karyotypes of all iPSC lines were normal (Supplementary Figure S1). Sendai virus was not detected in all iPSC lines (Supplementary Table S6). Pluripotency of iPSCs were examined by gene expression analysis (Supplementary Table S6), flow cytometry (Supplementary Figures S2A–D) and immunocytochemistry (Supplementary Figure S4A). The iPSC-derived NPCs were stained positive for neural progenitor markers Nestin and Musashi1 as well as neuronal marker Class III β-tubulin by immunocytochemistry (Supplementary Figure S4B). Flow cytometry also showed that the majority of NPCs expressed markers Nestin and Sox2 (Supplementary Figure S3). For generation of D30 neurons, the NPCs were cultured in NDM for 30 days. The D30 neurons expressed neuronal markers MAP2 and Class III β-tubulin (Supplementary Figure S5F). Western blot detected synaptic vesicle protein synapsin I (SYN1), which regulates neurotransmitter release, in D30 neurons (Supplementary Figure S5G). To characterize the type(s) of neurons in D30 cultures, we stained for glutamatergic neuron marker VGLUT1, GABAergic neuron marker VGAT and glia cell marker GFAP (Supplementary Figure S4C). Immunocytochemistry showed subpopulations of neurons stained positive for VGLUT1, VGAT or GFAP, indicating heterogeneity. We also examined the calcium influx of the D30 neurons from the Uaf, TD1 and TD2 subjects. All three exhibited spontaneous calcium influx and potassium-stimulated calcium influx when depolarized with the 100 mM KCl Tyrode’s solution (Supplementary Figures S5A–E), further indicating that the iPSC-derived D30 neurons have appropriate neuronal phenotypes.
To explore whether neurons derived from iPSCs of controls as well as TD patients have properties resembling mature neurons, we performed patch-clamp recordings of neurons at 30–37 days after replating the 15 days of neurons on glial cells. These neurons expressed neuronal markers MAP2, Class III β tubulin (Figure 2a). Under voltage-clamp mode we observed both fast, inactivating inward and outward currents, which probably correspond to opening of voltage-dependent Na+ and K+ -channels, respectively (Figure 2b). More importantly, a large proportion of these neurons can fire spontaneous action potentials (Figure 2c). We also conducted IHCs and revealed that synapsin punctas were found in these cultures (Figure 2d). The basic membrane properties of these human neurons were comparable among different cell lines (Figures 2e–g).
Functional characterizations of neurons derived from iPSCs. (a) Neurons derived from unaffected as wells as TD subjects express mature neuronal marker MAP2 and Class III β-tublin. (b) Whole cell currents responses were recorded from human neurons derived from subjects with and without TD. (c) Spontaneous action potentials were recorded from human neurons derived from subjects with and without TD. (d) Synapsin, a synaptic vesicle marker, was detected in human neurons with and without TD. (e) Membrane capacitance, (f) Membrane resistance and (g) resting membrane potentials of human neurons derived from subjects with and without TD were quantified. iPSC, induced pluripotent stem cells; TD, Tourette disorder.
The human PNKD (L) isoform is enriched in iPSC-derived neurons and is reduced in TD patients’ neurons
The 12 exons within the human PNKD locus encode multiple protein isoforms.17 The PNKD (L) transcript contains exons 1, 2 and 5–12. The PNKD medium (M) transcript contains exons 4–12 and the PNKD short (S) transcript contains exons 1–3. The nonsense mutation is in exon 6, therefore affecting both the PNKD (L) and PNKD (M) transcripts. The Genotyping-Tissue Expression (GTEx) project (http://www.gtexportal.org/home/) indicates that PNKD (L) transcript is enriched in the human brain where PNKD (M) transcript is weakly expressed. The PNKD short (S) transcript has higher expression than the long transcript in the human brain but the PNKD (S) transcript is also highly expressed in other human tissues. To characterize PNKD gene expression, all PNKD transcripts were quantitated by RT–qPCR in lymphoblastoid cell lines (LCL),7 iPSC, NPC and D30 neurons from subjects Uaf, TD1 and TD2. Primers distinguished different transcripts and their positions are shown in Supplementary Table S1. In Figure 3, we show PNKD transcript expression in the Uaf subject. The PNKD (L) transcript was not detected in LCL and iPSC but appeared at low levels in NPC, whereas its expression increased greatly in the D30 neurons (Figure 3a). PNKD (L) transcript expression significantly increased from iPSC to NPC and from NPC to D30 neurons (Figure 3a). The PNKD (S) transcript was ubiquitously detected with relatively higher expression and it increased significantly with neuronal differentiation of iPSC (Figure 3b). The PNKD (M) transcript is expressed by all cell types examined, but its expression was relatively low in neuronal cells (Figure 3c). PNKD (L) protein expression was examined by western blot in LCL, iPSC and D30 neurons of all three subjects. Consistent with the transcript expression data, the PNKD (L) protein was only detected in D30 neurons (Figure 3e) and Urel_Unaf (Supplementary Figure S6A).Thus, of the two PNKD isoform transcripts affected by the nonsense mutation, the (L) but not the (M) isoform is enriched in human neuronal cells. Nonsense mutations could produce loss-of-function, gain-of-function or dominant-negative phenotypes. To investigate effects of the nonsense mutation on the PNKD (L) isoform, we measured the transcript levels and protein expression in D30 neurons. For each subject, D30 neurons from three independent differentiations were included in the RT–qPCR analysis. The PNKD (L) transcript levels in the two TD patients’ neurons were reduced to about half of the level of the Uaf subject’s neurons, suggesting nonsense-mediated decay (NMD)18 (Figure 3d). Consistent with the transcript expression, the PNKD (L) protein was also reduced in the TD patients’ neurons compared to the Uaf (Figure 3e).
PNKD expression in LCL, NPC, iPSC and iPSC-derived D30 neurons by RT–qPCR. (a) PNKD (L) transcript expression increased from iPSC to NPC and from NPC to D30 neurons in the Uaf subject, indicating that PNKD (L) was enriched in neuronal cells (n=3). (b) PNKD (S) transcript is expressed in all the cell types of all three subjects. PNKD (S) transcript level increased during neural differentiation (n=3). (c) PNKD (M) transcript is expressed at low levels in iPSCs, NPCs and iPSC-derived neurons (n=3). (d) PNKD (L) transcript was significantly reduced in iPSC-derived D30 neurons of TD1 and TD2 subjects compared to the Uaf subject (n=3). (e) PNKD (L) protein was detected in D30 neurons but not in LCL or iPSC. PNKD (L) protein was reduced in iPSC-derived D30 neurons of subjects TD1 and TD2 compared to the Uaf subject (n=3). iPSC, induced pluripotent stem cell; LCL, lymphoblastoid cell line; NPC, neural progenitor cell.
Furthermore, we have observed that treating TD neurons with 100 μg ml−1 NMD inhibitor Cycloheximide eliminated the expression difference of PNKD (L) transcript between unaffected and TD neurons (Supplementary Figure S6B), supporting our NMD hypothesis.
The human PNKD (L) protein self-oligomerizes and interacts with RIMS1α protein
A previous study showed that the human PNKD (L) protein interacts with itself.19 We transfected 293FT cells with plasmids expressing: GFP, FLAG-PNKD (L), PNKD (L)-Myc or FLAG-PNKD (L)+PNKD (L)-Myc. Forty-eight hours after transfection, cells were lysed for co-immunoprecipitation. The FLAG-PNKD (L) was eluted by anti-FLAG M2 magnetic beads. The PNKD (L)-Myc co-eluted with FLAG-PNKD (L), indicating the self-oligomerization of the PNKD (L) protein (Figure 4a). It has been reported that the PNKD (L) protein interacts with the presynaptic proteins RIMS1/2 in mouse cortex.20 Based on the mouse and human protein sequence conservation, the PNKD (L) and RIMS1α proteins also likely interact in human. We transfected the 293FT cells with plasmids expressing: GFP, RIMS1α-FLAG, PNKD (L)-Myc or RIMS1α-FLAG+PNKD (L)-Myc. co-immunoprecipitation was performed using the whole cell lysates and, as shown in Figure 4b, the PNKD (L)-Myc was eluted only in the presence of RIMS1α-FLAG, suggesting their interaction.
The human PNKD (L) protein oligomerizes with itself and interacts with the RIMS1α protein. (a) 239FT cells overexpressing GFP, FLAG-PNKD(L), PNKD(L)-Myc or FLAG-PNKD(L)+PNKD(L)-Myc were lysed for Co-IP. The PNKD(L)-Myc was pulled down by the anti-FLAG magnetic beads only in the presence of the FLAG-PNKD(L). (b) 239FT cells overexpressing GFP, RIMS1α-FLAG, PNKD(L)-Myc or RIMS1α-FLAG+PNKD(L)-Myc were lysed for Co-IP. The PNKD(L)-Myc was eluted by the anti-FLAG magnetic beads only in the presence of the RIMS1α-FLAG. Co-IP, co-immunoprecipitation.
We also examined the subcellular localization of PNKD (L) and RIMS1α in neurons. Owing to the low endogenous expression of PNKD (L), by transfection we overexpressed the PNKD (L) with the FLAG tag at its N-terminus in iPSC-derived neurons. We also transfected the iPSC-derived neurons with plasmid expressing the human RIMS1α protein with the Myc tag at its C-terminus. Immunocytochemistry showed that PNKD (L) protein was enriched in the plasma membrane and neurites whereas expression was weaker in the cell body (Supplementary Figure S7A). The RIMS1α protein could be detected in both cell body and neurites (Supplementary Figure S7B). Co-transfection of the PNKD (L) and the RIMS1α showed that these two proteins co-localized in the cell body and neurites (Supplementary Figure S7C).
RIMS1α transcript is enriched in iPSC-derived neurons and is not affected by the reduction of the PNKD (L) isoform
GTEx indicates that the RIMS1α transcript is exclusively expressed in the brain. Using RT–qPCR we measured the level of the RIMS1α transcript in LCL, iPSC, NPC and D30 neurons of Uaf, TD1 and TD2. As shown in Figure 5a, the RIMS1α transcript is barely detected in LCL and has extremely low expression in iPSC and NPC. As expected, the RIMS1α transcript is highly enriched in D30 neurons. We have shown that PNKD (L) and the RIMS1α protein interact in 293FT cells and co-localized in iPSC-derived neurons. Additionally, it has been shown that PNKD (L) stabilizes RIMS1/2 proteins in mouse.20 Therefore, we measured the endogenous RIMS1 protein in D30 neurons from three independent differentiation processes. As shown in Figure 5c, the RIMS1 protein was only reduced in TD1 and TD2 neurons, correlating with the reduction of the PNKD (L) protein. To investigate whether the reduction of PNKD (L) affects the expression of the RIMS1α transcript in human neurons, we measured the RIMS1α transcript in D30 neurons from Uaf, TD1 and TD2 (Figure 5b). There is no significant change of the RIMS1α transcript expression in Uaf or TD neurons, ruling out a transcriptional regulatory mechanism.
Endogenous RIMS1α expression in Uaf and TD cells. (a) RIMS1α transcript was not expressed in LCL, was expressed at low levels in iPSC and NPC and was enriched in iPSC-derived D30 neurons (n=3). (b) RT–qPCR measured the RIMS1α transcript in iPSC-derived D30 neurons of Uaf, TD1 and TD2 subjects. D30 neurons from three independent differentiations were collected. There was no significant difference between the expression of the RIMS1α transcript in Uaf and TD neurons (n=3). (c) RIMS1 protein was examined by western blot in D30 neurons from three independent differentiations. RIMS1 protein was reduced in TD neurons. iPSC, iPSC, induced pluripotent stem cell; LCL, lymphoblastoid cell lines; NPC, neural progenitor cell; TD, Tourette disorder.
Discussion
Expression of the PNKD and RIMS1 in the human brain
BrainSpan (http://www.brainspan.org/) developmental transcriptome data show that PNKD gene expression (RPKM) increases with human brain development (Supplementary Figure S8A). We also searched PNKD expression in developing brains by isoform. Supplementary Figure S8B shows the expression of an exon only included in the PNKD (S) transcript, which has peaks at 6 months and at 4 years of age. Supplementary Figure S8C shows the expression of an exon only included in the PNKD (M) transcript. By comparing the RPKM values, the expression of the PNKD (M) transcript at all ages is much lower and not coordinated with the PNKD (S) transcript. Unfortunately, there is no exon that is specific to the PNKD (L) transcript. Supplementary Figure S8D shows the expression of an exon shared by the PNKD (L) and the PNKD (M) transcripts, which increases greatly during human brain development. The increase is probably due to the increased expression of the PNKD (L) transcript considering the uniformly low expression of the PNKD (M) transcript. Even though 26 brain structures are listed in Supplementary Figure S8, not every human subject could provide all 26 samples. Still, for the exon shared by PNKD (L) and PNKD (M) transcripts, we listed the brain structure which shows the highest expression in each subject (Supplementary Table S5). Interestingly, brain structures implicated in the neuropathogenesis of TD, including striatum (STR),21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32 primary motor cortex (area M1, area 4) (M1C),33, 34, 35, 36, 37, 38, 39, 40, 41 primary somatosensory cortex (area S1, areas 3,1,2) (S1C)32, 34, 39, 42, 43 and mediodorsal nucleus of thalamus (MD)44 have the highest expression at certain stages during brain development, suggesting that disruption of PNKD (L) protein function might affect neuronal development or activity in these brain structures. We also plotted RIMS1 gene expression using gene-level reads (Supplementary Figure S9A) and RIMS1α expression using exon-level reads (Supplementary Figure S9B) using the RNA-Seq data from the BrainSpan. Both the RIMS1 gene total expression and the RIMS1α expression are enriched in the cerebellar cortex region in some of the subjects. Additionally, we examined the PNKD and RIMS1 expression in the Human Brain Transcriptome database (http://hbatlas.org/) where gene expression was measured by microarray at exon level using over 1340 samples from 57 brains. PNKD expression is shown in Supplementary Figure S10A, where an increasing trend is seen with human brain maturation. Also, PNKD might have higher expression in the striatum than in other brain regions. For RIMS1, an increase of expression was seen from period 1 to period 6 (Supplementary Figure S10B). RIMS1 expression reached a plateau for mediodorsal nucleus of the thalamus (MD), striatum (STR), amygdala,45 hippocampus46 and 11 areas of neocortex (NCX), except for the cerebellar cortex. The cerebellar cortex has higher expression of RIMS1 from period 8 to period 15 compared to other brain regions. We modeled the expression of PNKD transcript isoforms during very early brain development by measuring PNKD (L), the PNKD (M) and the PNKD (S) transcripts in human iPSC, NPCs and D30 neurons (Figure 3). Consistent with brain observations, both the PNKD (L) and the PNKD (S) transcripts increased during neural differentiation, suggesting these two isoforms have neuronal functions, especially the former as its increase is tissue-specific and most pronounced.
Effect of the PNKD nonsense mutation
We observed reduced PNKD (L) transcript and protein levels in the mutant D30 neurons, suggestive of NMD. We investigated a possible dominant-negative mechanism for reduction of PNKD multimer activity due to a putative N-terminal PNKD protein fragment binding to full length fragment(s). Therefore, we determined whether NMD was complete by measuring PNKD (L) transcript with the mutation in iPSC-derived neurons. We reverse-transcribed total RNA extracted from Uaf and TD iPSC-derived neurons into cDNA and used the cDNA as template to PCR-amplify the region containing the nonsense variant. The forward and reverse PCR primers located at different exons. In addition, the reverse primer spanned an exon–exon junction. Therefore, only cDNA and not genomic DNA could be amplified. The PCR product was purified from the agarose gel and Sanger sequenced. As shown in Supplementary Figure S11A, the PNKD (L) transcript carrying the nonsense variant was detected, indicating that the NMD does not eliminate all the mutant PNKD (L) transcripts. However, we were unable to determine whether the mutant PNKD (L) transcript is translated into truncated protein. To get an idea of the possible subcellular localization of putative PNKD truncated protein, we overexpressed the mutant PNKD (L) transcript in 293FT cells where it was translated into a truncated protein. As shown in Supplementary Figure S11B, the truncated PNKD protein lost the ability to localize to the plasma membrane and did not co-localize with the wild-type PNKD (L) protein, arguing against a dominant-negative effect. Therefore, we hypothesize that the nonsense mutation exerts its phenotype due to haploinsufficiency at critical times in development. However, in certain brain neurons that express the PNKD (L) transcript at a much higher level, testing for a truncated PNKD (L) protein might be useful.
The PNKD gene and disease
The PNKD gene is known for its association with the neurological movement disorder PNKD which causes episodic involuntary movement attacks.14, 15, 16 As shown in Table 1, recurrent missense mutations A7V15, 47, 48, 49, 50, 51, 52, 53, 54 and A9V15, 48, 49, 50, 55, 56, 57 were associated with the PNKD disorder in familial cases from different ethnic groups. Transgenic mice expressing Pnkd carrying the human disease-causing mutations A7V and A9V exhibited alterations of striatal dopamine signaling.58 Another heterozygous mutation A33P17 was detected in one patient who suffered PNKD and cerebellar ataxia. Also a missense mutation G89R was identified in a boy affected by intermittent ataxia, diarrhea, exercise intolerance and speech articulation problems.59 In addition to missense mutations, a single nucleotide deletion (P341Rfs*2) in the PNKD (L) transcript leading to an amino acid change at position 341 (Proline to Arginine) and the generation of a premature stop codon at position 343 was reported to co-segregate with familial hemiplegic migraine.60 Mutations affecting the PNKD (L) and PNKD (S) isoforms including A7V, A9V, A33P were associated with the classic, autosomal dominant PNKD whereas mutations affecting the PNKD (L) and PNKD (M) isoforms including the G89R and P341Rfs*2 were associated with ataxia or familial hemiplegic migraine. It has been reported that episodic ataxia type-2 and familial hemiplegic migraine could be caused the by mutations in the same gene: CACNL1A4.61 Our TD and Tic disorder-associated PNKD nonsense mutation affects the PNKD (L) and the PNKD (M) isoforms leading to reduction of PNKD (L) in the TD neurons. Longer than the truncated protein caused by our nonsense mutation, the truncated protein due to the P341Rfs*2 mutation (60) presumably preserves the region that was required for PNKD (L) self-oligomerization19 and therefore it might interfere with the function of PNKD (L) protein. Interestingly, previous studies suggested clinical overlap between migraine and TD62, 63 or between paroxysmal kinesigenic dyskinesia (PKD, an involuntary movement distinct from PNKD disorder) and TD in multiple cases.64, 65, 66 Taken together, disorders like PNKD, PKD, familial hemiplegic migraine, TD and Tic disorders, whose diagnostic criteria and clinical phenotypes differ, may be caused by distinct mutations disrupting the same gene.
To summarize, we identified a nonsense mutation in the PNKD gene segregating with TD and Tic disorders in a multiplex pedigree. We characterized the functional consequences of the mutation in iPSC-derived neurons and demonstrated the self-oligomerization of the PNKD (L) protein and its interaction with the synaptic active zone protein RIMS1α. In addition, we analyzed the spatial and temporal expression of PNKD in publicly available human brain transcriptomic data. Also, we discuss the data-supported alternatives of haploinsufficiency versus a dominant-negative phenotypic mechanism. This report increases the evidence that PNKD plays a critical role in neuronal development and function but further studies will be required to fully elucidate the role of the PNKD gene in the pathogenesis of neurodevelopmental disorders, including TD.
References
Sun N, Tischfield JA, King RA, Heiman GA . Functional evaluations of genes disrupted in patients with Tourette’s disorder. Front Psychiatry 2016; 7: 11.
Pauls DL, Fernandez TV, Mathews CA, State MW, Scharf JM . The inheritance of Tourette disorder: a review. J Obsessive Compuls Relat Disord 2014; 3: 380–385.
Scharf JM, Yu D, Mathews CA, Neale BM, Stewart SE, Fagerness JA et al. Genome-wide association study of Tourette's syndrome. Mol Psychiatry 2013; 18: 721–728.
Ercan-Sencicek AG, Stillman AA, Ghosh AK, Bilguvar K, O'Roak BJ, Mason CE et al. L-histidine decarboxylase and Tourette's syndrome. N Engl J Med 2010; 362: 1901–1908.
Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science 2005; 310: 317–320.
Dietrich A, Fernandez TV, King RA, State MW, Tischfield JA, Hoekstra PJ et al. The Tourette International Collaborative Genetics (TIC Genetics) study, finding the genes causing Tourette syndrome: objectives and methods. Eur Child Adolesc Psychiatry 2015; 24: 141–151.
Heiman GA, King RA, Tischfield JA . New Jersey Center for Tourette Syndrome sharing repository: methods and sample description. BMC Med Genomics 2008; 1: 58.
Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat Biotechnol 2014; 32: 663–669.
Jennifer CM, Michael HS, Ronald PH . Biobanking in the Era of the Stem Cell: A Technical and Operational Guide. Morgan & Claypool Life Sciences, 2012 p 82.
Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M . Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010; 463: 1035–1041.
Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ et al. Induction of human neuronal cells by defined transcription factors. Nature 2011; 476: 220–223.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291.
Willsey AJ, Fernandez TV, Yu D, King RA, Dietrich A, Xing J et al. De novo coding variants are strongly associated with tourette disorder. Neuron 2017; 94: 486–499 e489.
Fouad GT, Servidei S, Durcan S, Bertini E, Ptacek LJ . A gene for familial paroxysmal dyskinesia (FPD1) maps to chromosome 2q. Am J Hum Genet 1996; 59: 135–139.
Lee HY, Xu Y, Huang Y, Ahn AH, Auburger GW, Pandolfo M et al. The gene for paroxysmal non-kinesigenic dyskinesia encodes an enzyme in a stress response pathway. Hum Mol Genet 2004; 13: 3161–3170.
Demirkiran M, Jankovic J . Paroxysmal dyskinesias: clinical features and classification. Ann Neurol 1995; 38: 571–579.
Ghezzi D, Viscomi C, Ferlini A, Gualandi F, Mereghetti P, DeGrandis D et al. Paroxysmal non-kinesigenic dyskinesia is caused by mutations of the MR-1 mitochondrial targeting sequence. Hum Mol Genet 2009; 18: 1058–1064.
Kervestin S, Jacobson A . NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 2012; 13: 700–712.
Shen Y, Lee HY, Rawson J, Ojha S, Babbitt P, Fu YH et al. Mutations in PNKD causing paroxysmal dyskinesia alters protein cleavage and stability. Hum Mol Genet 2011; 20: 2322–2332.
Shen Y, Ge WP, Li Y, Hirano A, Lee HY, Rohlmann A et al. Protein mutated in paroxysmal dyskinesia interacts with the active zone protein RIM and suppresses synaptic vesicle exocytosis. Proc Natl Acad Sci USA 2015; 112: 2935–2941.
Singer HS, Minzer K . Neurobiology of Tourette’s syndrome: concepts of neuroanatomic localization and neurochemical abnormalities. Brain Dev 2003; 25 (Suppl 1): S70–S84.
Kataoka Y, Kalanithi PS, Grantz H, Schwartz ML, Saper C, Leckman JF et al. Decreased number of parvalbumin and cholinergic interneurons in the striatum of individuals with Tourette syndrome. J comp neurol 2010; 518: 277–291.
Rapanelli M, Frick LR, Pogorelov V, Ota KT, Abbasi E, Ohtsu H et al. Dysregulated intracellular signaling in the striatum in a pathophysiologically grounded model of Tourette syndrome. Eur Neuropsychopharmacol 2014; 24: 1896–1906.
Israelashvili M, Bar-Gad I . Corticostriatal divergent function in determining the temporal and spatial properties of motor tics. J Neurosci 2015; 35: 16340–16351.
Pogorelov V, Xu M, Smith HR, Buchanan GF, Pittenger C . Corticostriatal interactions in the generation of tic-like behaviors after local striatal disinhibition. Exp Neurol 2015; 265: 122–128.
Xu M, Kobets A, Du JC, Lennington J, Li L, Banasr M et al. Targeted ablation of cholinergic interneurons in the dorsolateral striatum produces behavioral manifestations of Tourette syndrome. Proc Natl Acad Sci USA 2015; 112: 893–898.
Lennington JB, Coppola G, Kataoka-Sasaki Y, Fernandez TV, Palejev D, Li Y et al. Transcriptome analysis of the human striatum in Tourette syndrome. Biol Psychiatry 2016; 79: 372–382.
Leckman JF, Bloch MH, Smith ME, Larabi D, Hampson M . Neurobiological substrates of Tourette's disorder. J Child Adolesc Psychopharmacol 2010; 20: 237–247.
Kalanithi PS, Zheng W, Kataoka Y, DiFiglia M, Grantz H, Saper CB et al. Altered parvalbumin-positive neuron distribution in basal ganglia of individuals with Tourette syndrome. Proc Natl Acad Sci USA 2005; 102: 13307–13312.
Saka E, Graybiel AM . Pathophysiology of Tourette's syndrome: striatal pathways revisited. Brain Dev 2003; 25 (Suppl 1): S15–S19.
Peterson BS, Thomas P, Kane MJ, Scahill L, Zhang H, Bronen R et al. Basal Ganglia volumes in patients with Gilles de la Tourette syndrome. Arch Gen Psychiatry 2003; 60: 415–424.
Wang ZS, Maia TV, Marsh R, Colibazzi T, Gerber A, Peterson BS . The neural circuits that generate tics in Tourette's syndrome. Am J Psychiat 2011; 168: 1326–1337.
Ziemann U, Paulus W, Rothenberger A . Decreased motor inhibition in Tourette’s disorder: evidence from transcranial magnetic stimulation. Am J Psychiatry 1997; 154: 1277–1284.
Sowell ER, Kan E, Yoshii J, Thompson PM, Bansal R, Xu D et al. Thinning of sensorimotor cortices in children with Tourette syndrome. Nat Neurosci 2008; 11: 637–639.
Stinear CM, Coxon JP, Byblow WD . Primary motor cortex and movement prevention: where Stop meets Go. Neurosci Biobehav R 2009; 33: 662–673.
Franzkowiak S, Pollok B, Biermann-Ruben K, Sudmeyer M, Paszek J, Jonas M et al. Altered pattern of motor cortical activation-inhibition during voluntary movements in Tourette syndrome. Mov Disord 2010; 25: 1960–1966.
Suppa A, Belvisi D, Bologna M, Marsili L, Berardelli I, Moretti G et al. Abnormal cortical and brain stem plasticity in Gilles de la Tourette syndrome. Mov Disord 2011; 26: 1703–1710.
Franzkowiak S, Pollok B, Biermann-Ruben K, Sudmeyer M, Paszek J, Thomalla G et al. Motor-cortical interaction in Gilles de la Tourette syndrome. PLoS One 2012; 7: e27850.
Bunse T, Wobrock T, Strube W, Padberg F, Palm U, Falkai P et al. Motor cortical excitability assessed by transcranial magnetic stimulation in psychiatric disorders: a systematic review. Brain Stimul 2014; 7: 158–169.
Gilbert DL, Bansal AS, Sethuraman G, Sallee FR, Zhang J, Lipps T et al. Association of cortical disinhibition with tic, ADHD, and OCD severity in Tourette syndrome. Mov Disord 2004; 19: 416–425.
Berardelli A, Curra A, Fabbrini G, Gilio F, Manfredi M . Pathophysiology of tics and Tourette syndrome. J Neurol 2003; 250: 781–787.
Thomalla G, Siebner HR, Jonas M, Baumer T, Biermann-Ruben K, Hummel F et al. Structural changes in the somatosensory system correlate with tic severity in Gilles de la Tourette syndrome. Brain 2009; 132 (Pt 3): 765–777.
Fahim C, Yoon U, Das S, Lyttelton O, Chen J, Arnaoutelis R et al. Somatosensory-motor bodily representation cortical thinning in Tourette: effects of tic severity, age and gender. Cortex 2010; 46: 750–760.
Gilbert DL, Christian BT, Gelfand MJ, Shi B, Mantil J, Sallee FR . Altered mesolimbocortical and thalamic dopamine in Tourette syndrome. Neurology 2006; 67: 1695–1697.
Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014; 17: 1418–1428.
Banka S, Veeramachaneni R, Reardon W, Howard E, Bunstone S, Ragge N et al. How genetically heterogeneous is Kabuki syndrome?: MLL2 testing in 116 patients, review and analyses of mutation and phenotypic spectrum. Eur J Hum Genet 2012; 20: 381–388.
Rainier S, Thomas D, Tokarz D, Ming L, Bui M, Plein E et al. Myofibrillogenesis regulator 1 gene mutations cause paroxysmal dystonic choreoathetosis. Arch Neurol 2004; 61: 1025–1029.
Chen DH, Matsushita M, Rainier S, Meaney B, Tisch L, Feleke A et al. Presence of alanine-to-valine substitutions in myofibrillogenesis regulator 1 in paroxysmal nonkinesigenic dyskinesia: confirmation in 2 kindreds. Arch Neurol 2005; 62: 597–600.
Djarmati A, Svetel M, Momcilovic D, Kostic V, Klein C . Significance of recurrent mutations in the myofibrillogenesis regulator 1 gene. Arch Neurol 2005; 62: 1641.
Hempelmann A, Kumar S, Muralitharan S, Sander T . Myofibrillogenesis regulator 1 gene (MR-1) mutation in an Omani family with paroxysmal nonkinesigenic dyskinesia. Neurosci Lett 2006; 402: 118–120.
Yeh TH, Lin JJ, Lai SC, Wu-Chou YH, Chen AC, Yueh KC et al. Familial paroxysmal nonkinesigenic dyskinesia: clinical and genetic analysis of a Taiwanese family. J Neurol Sci 2012; 323: 80–84.
Fekete R . Paroxysmal nonkinesigenic dyskinesia with tremor. Case Rep Neurol Med 2013; 2013: 927587.
Pons R, Cuenca-Leon E, Miravet E, Pons M, Xaidara A, Youroukos S et al. Paroxysmal non-kinesigenic dyskinesia due to a PNKD recurrent mutation: report of two Southern European families. Eur J Paediatr Neuro 2012; 16: 86–89.
Liang SL, Yu XM, Zhang SH, Tai JL . A case of familial paroxysmal nonkinesigenic dyskinesia due to mutation of the PNKD gene in Chinese Mainland. Brain Res 2015; 1595: 120–126.
Stefanova E, Djarmati A, Momcilovic D, Dragasevic N, Svetel M, Klein C et al. Clinical characteristics of paroxysmal nonkinesigenic dyskinesia in Serbian family with Myofibrillogenesis regulator 1 gene mutation. Mov Disord 2006; 21: 2010–2015.
Ghezzi D, Canavese C, Kovacevic G, Zamurovic D, Barzaghi C, Giorgi C et al. A family with paroxysmal nonkinesigenic dyskinesias (PNKD): evidence of mitochondrial dysfunction. Eur J Paediatr Neurol 2015; 19: 64–68.
Szczaluba K, Jurek M, Szczepanik E, Friedman A, Milewski M, Bal J et al. A family with paroxysmal nonkinesigenic dyskinesia: genetic and treatment issues. Pediatr Neurol 2009; 41: 135–138.
Lee HY, Nakayama J, Xu Y, Fan X, Karouani M, Shen Y et al. Dopamine dysregulation in a mouse model of paroxysmal nonkinesigenic dyskinesia. J Clin Invest 2012; 122: 507–518.
Courtagen Diagnostics Laboratory. nucSEEK Case Report 2013.
Gardiner AR, Jaffer F, Dale RC, Labrum R, Erro R, Meyer E et al. The clinical and genetic heterogeneity of paroxysmal dyskinesias. Brain 2015; 138 (Pt 12): 3567–3580.
Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 1996; 87: 543–552.
Kwak C, Vuong KD, Jankovic J . Migraine headache in patients with Tourette syndrome. Arch Neurol 2003; 60: 1595–1598.
Barabas G, Matthews WS, Ferrari M . Tourette's syndrome and migraine. Arch Neurol 1984; 41: 871–872.
Oyama G, Okun MS, Ashizawa T, Malaty IA . Paroxysmal kinesigenic dyskinesia-like symptoms in a patient with tourette syndrome. Tremor Other Hyperkinet Mov (N Y) 2011; 1.
Jan JE, Freeman RD, Good WV . Familial paroxysmal kinesigenic choreo-athetosis in a child with visual hallucinations and obsessive-compulsive behaviour. Dev Med Child Neurol 1995; 37: 366–369.
Bruno MK, Hallett M, Gwinn-Hardy K, Sorensen B, Considine E, Tucker S et al. Clinical evaluation of idiopathic paroxysmal kinesigenic dyskinesia: new diagnostic criteria. Neurology 2004; 63: 2280–2287.
Acknowledgements
We thank the family who have participated in and contributed to this study. We also thank the NIMH Repository and Genomics Resource (U24MH068457 to JAT) at RUCDR Infinite Biologics for transforming cell lines and for whole exome sequencing. We are very grateful to Dr Louis Ptacek at UCSF, who kindly sent us the anti-PNKD (L) (C-terminus) antibody. This study was supported by grants from the National Institute of Mental Health (R01MH092293 to GAH and JAT) and the New Jersey Center for Tourette Syndrome and Associated Disorders (NJCTS; to GAH and JAT). We are also grateful to the NJCTS for facilitating the inception and organization of the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Sun, N., Nasello, C., Deng, L. et al. The PNKD gene is associated with Tourette Disorder or Tic disorder in a multiplex family. Mol Psychiatry 23, 1487–1495 (2018). https://doi.org/10.1038/mp.2017.179
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2017.179
This article is cited by
-
Genetic updates on paroxysmal dyskinesias
Journal of Neural Transmission (2021)
-
Integrated Mobile Element Scanning (ME-Scan) method for identifying multiple types of polymorphic mobile element insertions
Mobile DNA (2020)
-
European Multicentre Tics in Children Studies (EMTICS): protocol for two cohort studies to assess risk factors for tic onset and exacerbation in children and adolescents
European Child & Adolescent Psychiatry (2019)