Abstract
We conducted a genome-wide meta-analysis of cognitive empathy using the ‘Reading the Mind in the Eyes’ Test (Eyes Test) in 88,056 research volunteers of European Ancestry (44,574 females and 43,482 males) from 23andMe Inc., and an additional 1497 research volunteers of European Ancestry (891 females and 606 males) from the Brisbane Longitudinal Twin Study. We confirmed a female advantage on the Eyes Test (Cohen’s d=0.21, P<2.2 × 10−16), and identified a locus in 3p26.1 that is associated with scores on the Eyes Test in females (rs7641347, Pmeta=1.58 × 10−8). Common single nucleotide polymorphisms explained 5.8% (95% CI: 4.5%–7.2%; P=1.00 × 10−17) of the total trait variance in both sexes, and we identified a twin heritability of 28% (95% CI: 13%–42%). Finally, we identified significant genetic correlation between the Eyes Test and anorexia nervosa, openness (NEO-Five Factor Inventory), and different measures of educational attainment and cognitive aptitude.
Similar content being viewed by others
Introduction
Cognitive empathy, defined as the ability to recognize what another person is thinking or feeling, and to predict their behaviour based on their mental states, is vital for interpersonal relationships, which in turn is a key contributor of wellbeing. Cognitive empathy is distinct from affective empathy, the latter of which is defined as the drive to respond to another’s mental states with an appropriate emotion.1, 2 Difficulties in cognitive empathy have been found in different psychiatric conditions, particularly autism.3 The dissociation between cognitive and affective empathy (the latter is often intact in autism, for example, whilst it is invariably impaired in antisocial personality disorder) suggests these have independent biological mechanisms.
Differences in cognitive empathy have been identified in individuals with psychiatric conditions such as autism,4 schizophrenia5, 6 and anorexia nervosa.7 This includes either elevated or reduced cognitive empathy in comparison to neurotypical controls, either of which can contribute to difficulties in social interactions and wellbeing.8 However, although such alterations in cognitive empathy in psychiatric conditions are well established, little is known about the genetic correlates of cognitive empathy. For example, it is unclear to what extent, differences in cognitive empathy are a genetic risk factor for developing various psychiatric conditions. Furthermore, as previous studies have often used self-report or performance tests, results from these studies may be influenced by the characteristics of the test and/or factors associated with the psychiatric conditions themselves. In sum, from previous studies, it is difficult to tease apart the genetic and non-genetic contributions to performance in cognitive empathy, and how these relate to various psychiatric conditions.
Here we investigate the genetic architecture of this aspect of social cognition using a well-validated test, the ‘Reading the Mind in the Eyes’ Test (Eyes Test). The Eyes Test is a brief online test where participants are shown photographs of the eye regions and have to identify the appropriate emotion or mental state they express.2 It has been widely used to investigate differences in cognitive empathy in a range of neuropsychiatric conditions including autism,4 schizophrenia,9 bipolar disorder,10 anorexia nervosa11 and major depressive disorder.12 The NIMH Research Domain Criteria (RDoC) lists the Eyes Test as one of several important tests for characterizing variation in social processes, under the category of Perception and Understanding of Others (http://1.usa.gov/1Qs6MdI).13 We conducted a genome-wide association meta-analysis of cognitive empathy in more than 89,000 individuals of European ancestry, and investigated both single nucleotide polymorphism (SNP)-based and twin-based heritabilities. We further conducted bivariate genetic correlation analyses for psychiatric conditions, psychological traits and brain volumes. We finally conducted gene-based enrichment analysis and investigate potential genetic sources of sex differences.
Materials and methods
Participants
23andMe
Research participants were customers of 23andMe, and have been described in detail elsewhere.14, 15 All participants completed an online version of the ‘Reading the Mind in the Eyes’ test (Eyes Test)2 online on the 23andMe research participant website (36 items). In total, 88,056 participants (44,574 females and 43,482 males) of European ancestry completed the Eyes Test and were genotyped. All participants provided informed consent and answered questions online according to 23andMe’s human subjects protocol, which was reviewed and approved by Ethical & Independent Review Services, an AAHRPP-accredited private institutional review board (http://www.eandireview.com). Only participants who were primarily of European ancestry (97% European Ancestry) were selected for the analysis using existing methods.16 Unrelated individuals were selected using a segmental identity-by-descent algorithm.17
Brisbane Longitudinal Twin Study
In addition, 1497 participants (891 females and 606 males) of Caucasian ancestry with genotype data from the Brisbane Longitudinal Twin Study (BLTS) completed the short version (14 questions) of the Eyes Test online as part of a study on genetic and environmental foundations of political and economic behaviors.18 Participant ages ranged from 18 to 73 (M=37, s.d.=14). Twin heritability was estimated from 749 twin individuals (including 122 complete monozygotic pairs and 176 complete dizygotic pairs). All participants provided online consent and the study was approved by the QIMR Berghofer Human Research Ethics Committee.
Measures
The ‘Reading the Mind in the Eyes’ Test (Eyes Test) is a brief questionnaire of cognitive empathy. Participants are shown scaled, black and white photographs of eye regions of actors and they have to choose the cognitive state portrayed from the four options provided. The Eyes Test has good test–retest reliability (for example, reliability of 0.833 in the Italian version,19 and 0.63 in the Spanish version20),4 and scores are unimodally and near-normally distributed in the general population. In the BLTS data set, there was a modest test–retest correlation of 0.47 in 259 participants who retook the test after a gap of nearly 2 years (Supplementary Note section 1). For each correct answer on the Eyes Test, participants score 1 point, so the scores ranged from 0 to 36 on the full version of the Eyes Test and 0–14 on the short version of the Eyes Test. Further details are provided in Supplementary Note section 1.
Genotyping, imputation and quality control
23andMe cohort
DNA extraction, genotyping, imputation and initial quality control were completed by 23andMe. Participants provided saliva samples, and DNA extraction and genotyping were performed by the National Genetic Institute. All participants were genotyped using one of four different platforms (V1, V2, V3 and V4). Briefly, the V1 and V2 chips were based on the Illumina Human Hap550+ BeadChip (560,000 SNPs), the V3 on the Illumina OmniExpress+ Beadchip (950,000 SNPs). The V4 had a fully customized array of approximately 570,000 SNPs. Across all platforms, a total of 1,030,430 SNPs were genotyped. For this analysis, we included only participants with a genotype call rate greater than 98.5%, and SNPs that passed the Hardy–Weinberg Equilibrium Test at P<10−20 and had a genotype rate>90%. In addition, SNPs present only on platform V1, Chromosome Y and mitochondrial SNPs were excluded due to small sample sizes and unreliable genotype calling respectively. Using trio-data, where available, SNPs that failed the parent–offspring transmission test were also excluded. Imputation was performed using Minimac221 using the March 2012 (v3) release of the 1000 Genomes Phase 1 reference haplotypes phased using Beagle22 (V3.3.1). Our analyses were restricted to SNPs that had a minor allele frequency of at least 1%, which left 9,955,952 SNPs after quality control. Genotyping, imputation and preliminary quality control were performed by 23andMe.
BLTS cohort
The BLTS participants were genotyped on Illumina HumanCoreExome-12 v1.0 or Human610-Quad v1.0 chips. These samples were genotyped in the context of a larger genome-wide association project. Genotype data were screened for genotyping quality (GenCall<0.7 from the Human610-Quad v1.0), individual and SNP call rates (<0.95 and <0.99 for exome markers on the HumanCoreExome-12 v1.0 chip), Hardy–Weinberg Equilibrium (P<10−6) and MAF (<0.01). The data were checked for non-European ancestry, pedigree, sex and Mendelian errors. Data from the two different chips were separately phased using SHAPEIT2 and imputed to the 1000 Genomes reference panel (Phase 1 v3) using Minimac3. After imputation SNPs with a MAF<0.05% were excluded, leaving 11,133,794 SNPs for analyses. We further excluded SNPs with imputation r2<0.6 for meta-analysis.
Statistical analyses
Association analyses
Linear regression for the 23andMe cohort was performed for the Eyes Test scores using age, sex and the first four ancestry principal components as covariates. For the sex-stratified analyses, sex was excluded as a covariate. The same regression model was used for the BLTS after accounting for relatedness using RAREMETALWORKER. Inverse variance weighted meta-analysis was performed using Metal.23 Post meta-analysis, we excluded SNPs that were only genotyped in the BLTS cohort due to the small sample size, but included SNPs that were only genotyped in the 23andMe cohort. LD pruning was performed using Plink24 with an r2 of 0.1. We calculated the variance explained by each individual SNP25 using the following formula:

where R2g|c/(1−R2c) is the proportion of variance explained by the SNP after accounting for the effects of the covariates, t is the t-statistic of the regression co-efficient, k is the number of covariates and n is the sample size. We corrected for winner’s curse using an FDR-based approach.26
Heritability and genetic correlation
We used the intercept from Linkage Disequilibrium Score Regression (LDSR) to calculate genomic inflation in the meta-analysis due to population stratification27 (https://github.com/bulik/ldsc). The intercept for the non-stratified genome-wide association study (GWAS) was 1.01 (0.006), for the males-only GWAS was 1.006 (0.006), and for the females-only GWAS was 1.005 (0.006). SNP heritability and genetic correlation were calculated using LDSR. Difference in heritability between males and females was tested using:

where Zdiff is the Z score for the difference in heritability for a trait, (h2males−h2females) is the difference SNP heritability estimate in males and females, and SE is the standard errors for the heritabilities. We calculated two-tailed P-values in R. We performed genetic correlation using summary GWAS data using LDSR. For all genetic correlation analyses, we used LD data from the north west European population as implemented in LDSR. Intercepts were not constrained in the analyses. We used Bonferroni correction to correct for multiple testing in the genetic correlation. We also provide the Benjamini–Hochberg adjusted q-values in Supplementary Table 5. We note that Bonferroni correction is likely to be conservative due to the reasonably high degree of phenotypic and genetic correlations between the traits tested. Further details about the samples used are provided in Supplementary Note section 2.
Twin heritability
Twin heritability was estimated from 749 twin individuals (including 122 complete monozygotic pairs and 176 complete dizygotic pairs) in the BLTS using full information maximum likelihood in OpenMx28 in R, which makes use of all available data. All twins completed the short version of the Eyes Test, and for those who completed the test twice only their first attempt was included in analyses. ADE, ACE, AE, CE and E models were fit to the data and fit indices compared to determine the best-fitting model. Standardized variance components are reported from the best-fitting model, the AE model and, for completeness, from the ADE and ACE models. Further details are given in Supplementary Note section 4.
Gene-based analyses and sex difference analyses
We used MetaXcan29 using tissue weights from the GTEx to perform gene-based analysis (https://github.com/hakyimlab/MetaXcan). MetaXcan uses summary statistics to perform gene-based association analyses. It incorporates eQTL data from the GTEx consortium to infer gene level expression based on the summary GWAS statistics provided. This can be used to identify tissue-specific gene expression for the trait of interest. Here, we performed gene-based analysis for the non-stratified GWAS meta-analysis for nine neural tissues: anterior cingulate cortex (BA24), caudate basal ganglia, cerebellar hemisphere, cerebellum, cortex, frontal cortex (BA9), hippocampus, hypothalamus, nucleus accumbens basal ganglia and putamen basal ganglia, using gene-expression regression coefficients for these tissues from the GTEx project. This is based on tissues from 73 to 103 individuals. We chose neural tissues as cognitive empathy can be assumed to be a neural phenotype. As MetaXcan predicts expression level from SNP information, we filtered out genes whose correlation with predicted models of expression was<0.01, as incorporated in MetaXcan. This steps helps guard against false positives, by removing genes whose expressions are poorly predicted by the model. We used an FDR-based correction to correct for all the tests run across all the tissues. Details of sex-difference analyses are provided in Supplementary Note section 3.
Data availability
Summary level data may be requested from 23andMe and received subject to 23andMe’s standard data transfer agreement. We have also provided summary statistics for the first 10,000 LD-pruned SNPS for the three GWAS analyses (males-only, females-only and non-stratified) as Supplementary Data.
Results
Heritability
In collaboration with 23andMe and the BLTS cohort, we conducted three separate genome-wide association study meta-analyses (GWAMAs) of the Eyes Test: a males-only GWAMA (n=44,088), a females-only GWAMA (n=45,465) and a non-stratified GWAMA (n=89,553). The study protocol is provided in Figure 1. All participants from the 23andMe cohort completed the full version of the Eyes Test online, comprising 36 questions (mean score=27.47±3.67), while participants from the BLTS cohort completed the short version of the Eyes Test (14 questions, mean=8.85±2.34) (Supplementary Note Section 1). Scores on the Eyes Test were significantly associated with age and sex in the 23andMe cohort (age: −0.026±0.0007; P<2.2 × 10−16, sex (females): 0.77±0.02; P<2.2 × 10−16). We used LDSR to calculate the heritability explained by all the SNPs in HapMap3 with minor allele frequency>5%. We identified a significant narrow sense heritability of 5.8% (95% CI: 4.5%–7.2%; P=1.00 × 10−17) in the non-stratified GWAMA. We calculated the twin heritability from 749 twin individuals (including 122 complete monozygotic pairs and 176 complete dizygotic pairs) in the BLTS. Heritability, from the best-fitting additive genes/unique environment (AE) model, was 28% (95% CI: 13%–42%) (Supplementary Note Section 4).
Schematic diagram of the study protocol. 88,056 research participants of European ancestry from 23andMe completed the full version of the Eyes Test and were genotyped. An additional 1,497 participants of European ancestry from the Brisbane Longitudinal Twin Study completed the short version (14 questions) of the Eyes Test and genotyped. Genome-wide association meta-analysis was performed on the combined cohort of 89,553 participants. Three separate meta-analyses were performed: males-only, females-only and non-stratified. Subsequently, functional enrichment and gene-based analysis was performed for the non-stratified GWAMA. SNP heritability and genetic correlation using LDSR was performed for the GWAMA. Sex differences were also investigated using the same data set. In parallel, twin heritability was calculated from 749 twin individuals from the Brisbane Longitudinal Twin Study who had completed the short version of the Eyes Test. GWAMA, genome-wide meta-analysis; GWAS, genome-wide association study; LDSR, Linkage Disequilibrium Score regression; SNP, single nucleotide polymorphism.
Genetic correlation
We next investigated how the non-stratified Eyes Test is genetically correlated to psychiatric conditions and specific psychological and cognitive traits for which summary GWAS data were available (Supplementary Table 5). After correcting for multiple testing (Bonferroni correction alpha=2.08 × 10−3), we identified significant positive genetic correlations between Eyes Test scores and the NEO-Five Factor Inventory measure of openness (rg=0.54±0.14; P=1.17 × 10−4).30 We also identified significant positive correlations with different measures of cognition and education: college years (rg=0.40±0.06; P=2.48 × 10−11),31 educational attainment (0.34±0.04; P=1.49 × 10−17),32 and childhood cognitive aptitude (calculated as Spearman’s g and is, hence, independent of word knowledge)33 (rg=0.34±0.10; P=1.2 × 10−3). In addition, we identified a significant positive genetic correlation between the Eyes Test scores and anorexia nervosa (rg=0.25±0.08; P=1.9 × 10−3) (Figure 2). We did not identify a significant genetic correlation between autism and scores on the Eyes Test.
Genetic correlations between the Eyes Test and psychiatric conditions, psychological traits and subcortical brain volumes. Genetic correlations and standard errors for the Eyes Test (GWAMA). Figures above the bars represent P-values. All P-values with P<0.05 provided. * represents significant genetic correlations after Bonferroni correction. Point estimate represents the genetic correlation, and the error bars represent the standard errors. BPD features is borderline personality disorder features, ICV is intracranial volume. We have removed the genetic correlation for agreeableness from this figure due to the high standard errors. The genetic correlations, standard errors and P-values for all traits including agreeableness are provided in Supplementary Table 5.
We also investigated if subcortical brain volumes are correlated with performance on the Eyes Test. We used data from the ENIGMA consortium for six subcortical regions and intracranial volume.25 We excluded the amygdala, even though it is relevant for social cognition, as the low heritability of the amygdala could not be accurately quantified using LDSR.23 None of the correlations were significant after Bonferroni correction. However, we identified nominally significant positive correlation between the Eyes Test scores and the volumes of the caudate nucleus25 (rg=0.24±0.09; P=9.25 × 10−3) and volume of the putamen (rg=0.21±0.08; P=1.33 × 10−2), which together form the dorsal striatum. All genetic correlations are provided in Supplementary Table 5.
We also investigated sex-stratified genetic correlations between the Eyes Test and educational attainment, the only relevant phenotype where we had access to sex-stratified data. We identified a modest, significant genetic correlation between educational attainment and the Eyes Test in the males-only data set: rg=0.23±0.05; P=2.6 × 10−5. We identified a higher, significant genetic correlation between educational attainment and Eyes Test in the females-only data set: rg=0.39±0.06; P=5.88 × 10−11. These results suggest that females share greater pleiotropy between general cognition and cognitive empathy than males, indicating different genetic mechanisms for the development of cognitive empathy.
Genome-wide association meta-analyses
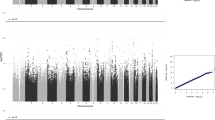
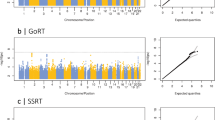
GWAMA of the non-stratified and the males-only data sets did not identify any significant loci. In the females-only analysis, we identified one locus at 3p26.2 that was significant at a threshold of P<5 × 10−8. This locus contains 21 significant SNPs in high LD (Figure 2), with concordant effect direction for 19 SNPs in the 23andMe and BLTS data sets. The leading SNP rs7641347 (Pmeta=1.58 × 10−8) explained 0.067% of the total variance, which reduced to 0.013% of the total variance after correcting for winner’s curse.26 Of the two SNPs with discordant effects in the two data sets, rs114076548 was the most-significant SNP in the 23andMe data set and had P=6.49 × 10−9. We did not identify any inflation in the P-values of the GWAMA due to population stratification using LDSR (intercept=1.01±0.007) (Figure 3).
Manhattan plot and regional association plot for the Eyes Test (females) meta-analysis GWAS. (a) Manhattan plot of the Eyes Test meta-analysis (female). x axis is the chromosomal position of the SNP, and y axis is the negative logarithm of the P-value. The red line indicates genome-wide significant threshold of 5 × 10−8. Lead SNP for all loci with P<1 × 10−6 is provided. n=44,574, and λgc=1.05. LDSR intercept=1.01. (b) Regional association plot of the significant locus for the Eyes Test (females) meta-analysis. GWAS, genome-wide association study; LDSR, Linkage Disequilibrium Score regression; SNP, single nucleotide polymorphism.
The leading SNP (rs7641347) is located in an intron of SUMF1 and was nominally significant in the non-stratified analysis (Pmeta=1.1 × 10−5), but non-significant in the males-only analysis (Pmeta=0.49). In addition, SNPs in high LD (r2>0.8) were also not nominally significant in the males-only analysis. Together, all 21 SNPs span a region of approximately 77 kb 3p26.2 (Supplementary Table 1). At this locus, in addition to SUMF1, two other genes are present: Leucine Rich Neuronal 1 (LRRN1) and SET Domain And Mariner Transposase Fusion Gene (SETMAR). LRRN1 is highly expressed in brain tissues,34 with highest median expression in the putamen, nucleus accumbens and the caudate nucleus, all three of which are part of the striatum. Deletion of 3p26.1 and 3p26.2 can cause developmental delay, hypotonia and epileptic seizures and has been implicated in autism.35
The most significant SNP in the males-only GWAS meta-analysis (rs4300633 in 16p12.3, P=9.11 × 10−8) explained 0.062% of the variance, and the most significant SNP in the non-stratified GWAS meta-analysis (rs149662397 in 17q21.32 P=1.58 × 10−7) explained only 0.029% of the variance. All LD-pruned SNPs in the three GWAMA analyses with P<1 × 10−6 are provided in Supplementary Table 2. The QQ-plot and locus-zoom plot for the females-only meta-analysis, and the Manhattan and QQ-plots for the males-only and non-stratified analyses are provided in Supplementary Note section 5. Gene-based analyses using MetaXcan29 for 10 neural tissues (Materials and methods) and functional enrichment analyses for the non-stratified GWAMA did not identify any significant results (Supplementary Tables 3 and 4).
Sex differences
We also investigated sex-differences in the Eyes Test. There was a significant female advantage on the scores of the full Eyes Test (males=27.08±3.75; females=27.85±3.55; cohen’s d=0.21, P<2 × 10−16), replicating previous results36 (Figure 4). There was no significant difference in males-only or females-only SNP heritability estimates (males=7.1%±1.1%, females=6.7%±1.1%; P=0.79). There was a reasonably high but incomplete genetic correlation between males and females (rg=0.68±0.12; P=2.70 × 10−8). Binomial sign test of LD-pruned nominally significant SNPs in the sex-stratified analyses identified that 61% (95% CI: 59–62%) of the SNPs had a concordant effect direction (P<2.2 × 10−16). We further investigated the effect direction and statistical significance of all independent SNPs with P<1 × 10−6. SNPs that were of suggestive significance in one sex were not nominally significant in the other (Supplementary Table 2 and Supplementary Note section 5), which was supported by Cochran’s Q-value. However, the effect sizes of these SNPs are likely to be inflated by winner’s curse, and after correcting for winner’s curse, we did not identify significant Cochran’s Q-value (Supplementary Table 2). We do note that the correction is conservative. Using MetaXcan29 we identified the top cortically expressed genes (P<0.05) for both sexes and calculated the overlap in the genes. We did not find any enrichment in gene overlap (Fold difference=1.2, P=0.264). We also investigated if there was an enrichment of female-overexpressed or male-overexpressed cortical genes for the Eyes Test (Materials and methods) and did not find any significant enrichment (Supplementary Note section 3 and Supplementary Tables 6 and 7).
Mean scores and SNP heritability. (a) Mean phenotypic scores and standard deviations for the Eyes Test in the 23andMe cohort. Point estimate provides the mean score, and the error bars represent standard deviations. Difference in mean scores between males and females was highly significant (P<2.2 × 10−6; Cohen’s d=0.21). Numbers in brackets indicate the number of participants in each GWAS. All: non-stratified GWAS; Females: Females-only GWAS; Males: Males-only GWAS. (b) Mean SNP heritability estimates and standard errors for the Eyes Test in the GWAMA. Point estimate provides mean SNP heritability, and error bar represents standard errors. There was no significant difference in SNP heritability estimates between males and females (P=0.79). Numbers in brackets indicate the number of participants in each GWAMA. All: non-stratified GWAMA; Females: Females-only GWAMA; Males: Males-only GWAMA. GWAMA, genome-wide association study meta-analyses; GWAS, genome-wide association study; SNP, single nucleotide polymorphism.
Discussion
To the best of our knowledge, this is the first large-scale genetic study investigating the genetic architecture of cognitive empathy. We investigated heritability estimates of the Eyes Test in two samples. In our sample of 749 twin individuals (which included 122 complete monozygotic pairs and 176 complete dizygotic pairs), heritability was approximately 28% (95% CI: 13%–42%). This is in keeping with previous studies that have investigated heritability of other facets of empathy in twins. A meta-analysis of empathy in twins identified that approximately a third of the variance is heritable.37 In our sample of 88,056 unrelated research volunteers from 23andMe, SNP-based heritability was estimated using LDSR, and approximately 5% of the trait was additively heritable. It is likely that heritability of cognitive empathy changes with age (which was significantly correlated with scores on the Eyes Test in this data set), as is observed in prosocial behavior.37 In our analyses, age was included as a covariate, and thus our SNP heritability estimate is likely to represent the lower bound of the SNP heritability.
We identified significant positive genetic correlations with different measures of cognitive ability including educational attainment. This reflects the phenotypic correlation between measures of cognitive empathy and cognitive ability. A meta-analysis has identified a significant positive correlation between scores on the Eyes Test and IQ (n=3583; r=0.24; 95% CI: 0.16–0.32),38 perhaps reflecting that the Eyes Test has a verbal component that includes a varied mental state lexicon (matching a mental state word to an emotional expression). Other tests of theory of mind are also positively correlated with cognitive aptitude and measures of intelligence.39, 40, 41 This may reflect that theory of mind and in particular joint attention in infancy may facilitate language development and learning from others.42 Theory of mind may also be related to cognitive aptitude and IQ because we often infer another person’s mental state through their speech – speech is the ‘print-out’ of a person’s mind – so verbal IQ and language skills may facilitate theory of mind and vice versa. We also found a significant positive genetic correlation with the NEO-Openness to experience which likely reflects a previous correlation at a phenotypic level between measures of empathy and personality.43 With psychiatric conditions, there was a significant positive correlation with anorexia. One study identified that individuals with anorexia report higher personal distress,44 a subscale on a widely used measure of empathy, while other studies have reported that deficits in social cognition in anorexia may be attributable to comorbid alexithymia.45 Our research suggests that a genetic contribution to higher cognitive empathy increases one’s genetic risk for anorexia. This warrants further research.
We did not identify a significant genetic correlation between the Eyes Test and autism. This may be due to heterogeneity in performance in the Eyes Test, since only a subset of individuals with autism show impaired performance on the Eyes Test.4, 13 In addition, the cognitive phenotype of autism involves non-social aspects (such as excellent attention to detail), not just social deficits. A meta-analysis reported global or selective deficits in performance on the Eyes Test in individuals with schizophrenia, anorexia, bipolar disorder and clinical depression, but preserved or even enhanced performance for individuals with non-clinical depression and borderline personality disorder.46 However, these studies are typically conducted in small sample sizes. Performance on the Eyes Test may be influenced by multiple factors related to psychiatric conditions, and may not measure a direct causal relationship between psychiatric conditions and cognitive empathy.
We also note the nominally significant genetic correlation between volume of the caudate nucleus, putamen and scores on the Eyes Test. Although the correlations were not significant after Bonferroni correction, this is of potential interest as neuroimaging studies have reported activation in both the putamen47 and caudate nucleus48 during tasks of social cognition. In humans, the ventral striatum is composed of the nucleus accumbens and olfactory tubercle, whereas the dorsal striatum is composed of the caudate nucleus and putamen. There is some evidence to support the role of the striatum in theory of mind,49 and further research is needed to confirm that cognitive and affective empathy utilize different neural circuits. Using larger GWAS samples of subcortical brain volumes will help better understand if common genetic variants contribute to both volumes of the dorsal striatum and cognitive empathy.
We also identified one locus that is significantly associated with empathy in females. The top SNP (rs7641347) had a P-value=1.58 × 10−8. One of the closest genes to this locus, LRRN1, is highly expressed in striatum according to the GTEx database. However, we were unable to identify any eQTL that specifically linked this locus to the gene. LRRN1 is a gene that is not well characterized. In chicks, Lrrn1 is necessary for the formation of the mid-brain hind-brain boundary.50 The locus was significant in females, nominally significant in the non-stratified analyses, and non-significant in the males-only analyses, suggesting a sex-specific involvement of this locus in cognitive empathy measured using the Eyes Test. We note that even with approximately 90,000 individuals this GWAMA was underpowered to detect loci of significant effect, owing to the relatively low variance explained per SNP. Future research needs to investigate the functional significance of LRRN1 in human brain development and its role in neurodevelopmental conditions.
It is also interesting to note that while twin and SNP-based heritability did not vary between the sexes in our study, we replicated the female-advantage on the Eyes Test in the largest sample to date. Sex-stratified analyses also allowed us to investigate the genetic correlation between males and females, and subsequently, sex-specific imputed gene expression in cortical tissues. Male-female genetic correlation was only modest, which was supported by a binomial sign test. In comparison, other traits for which we had sex-stratified data, genetic correlation was considerably higher (for example, self-reported empathy:51 rg=0.82±0.16, systemizing51: rg=1.0±0.16; educational attainment32: rg=0.91±0.02). We also did not identify a significant overlap between the genes identified for the sex-stratified GWAS. All of this suggests that there is some sex specificity in the genetic architecture of cognitive empathy. Investigating how this sex-specific architecture is expressed and interacts with prenatal steroid hormones52 will help shed further light on the biological contributions to the female superiority on the Eyes Test.
In conclusion, we identify a genetic locus that is associated with scores on the Eyes Test in females. We identify significant positive genetic correlations between scores on the Eyes Test and three phenotypes: anorexia nervosa, cognitive aptitude and educational attainment, and openness to experience. Phenotypic sex-differences for the Eyes Test may be partly due to different genetic architectures in males and females, interacting with postnatal social experience.
References
Baron-Cohen S, Chakrabart B . Understanding the genetics of empathy and the autistic spectrum. In: Baron-Cohen S, Tager-Flusberg H, Lombardo MV (eds). Understanding Other Minds: Perspectives from Developmental Social Neuroscience. Oxford University Press: Oxford, 2013.
Baron-Cohen S, Wheelwright S, Hill J, Raste Y, Plumb I . The ‘Reading the Mind in the Eyes’ Test revised version: a study with normal adults, and adults with Asperger syndrome or high-functioning autism. J Child Psychol Psychiatry 2001; 42: 241–251.
Decety J, Moriguchi Y . The empathic brain and its dysfunction in psychiatric populations: implications for intervention across different clinical conditions. Biopsychosoc Med 2007; 1: 22.
Baron-Cohen S, Bowen DC, Holt RJ, Allison C, Auyeung B, Lombardo MV et al. The ‘Reading the Mind in the Eyes’ test: complete absence of typical sex difference in ~400 men and women with autism. PLoS One 2015; 10: e0136521.
Bora E, Gökçen S, Veznedaroglu B . Empathic abilities in people with schizophrenia. Psychiatry Res 2008; 160: 23–29.
Michaels TM, Horan WP, Ginger EJ, Martinovich Z, Pinkham AE, Smith MJ . Cognitive empathy contributes to poor social functioning in schizophrenia: evidence from a new self-report measure of cognitive and affective empathy. Psychiatry Res 2014; 220: 803–810.
Calderoni S, Fantozzi P, Maestro S, Brunori E, Narzisi A, Balboni G et al. Selective cognitive empathy deficit in adolescents with restrictive anorexia nervosa. Neuropsychiatr Dis Treat 2013; 9: 1583–1589.
Tone EB, Tully EC . Empathy as a ‘risky strength’: a multilevel examination of empathy and risk for internalizing disorders. Dev Psychopathol 2014; 26: 1547–1565.
Lam BYH, Raine A, Lee TMC . The relationship between neurocognition and symptomatology in people with schizophrenia: social cognition as the mediator. BMC Psychiatr 2014; 14: 138.
Cusi AM, Macqueen GM, McKinnon MC . Patients with bipolar disorder show impaired performance on complex tests of social cognition. Psychiatry Res 2012; 200: 258–264.
Tapajóz Pereira de Sampaio F, Soneira S, Aulicino A, Allegri RF . Theory of mind in eating disorders and their relationship to clinical profile. Eur Eat Disord Rev 2013; 21: 479–487.
Berlim MT, McGirr A, Beaulieu M-M, Turecki G . Theory of mind in subjects with major depressive disorder: is it influenced by repetitive transcranial magnetic stimulation? World J Biol Psychiatry 2012; 13: 474–479.
Lombardo MV, Lai M-C, Auyeung B, Holt RJ, Allison C, Smith P et al. Unsupervised data-driven stratification of mentalizing heterogeneity in autism. Sci Rep 2016; 6: 35333.
Tung JY, Do CB, Hinds DA, Kiefer AK, Macpherson JM, Chowdry AB et al. Efficient replication of over 180 genetic associations with self-reported medical data. PLoS One 2011; 6: e23473.
Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, Francke U et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet 2011; 7: e1002141.
Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, Mountain JL et al. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS One 2012; 7: e34442.
Henn BM, Hon L, Macpherson JM, Eriksson N, Saxonov S, Pe’er I et al. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS One 2012; 7: e34267.
Hatemi PK, Smith K, Alford JR, Martin NG, Hibbing JR . The genetic and environmental foundations of political, psychological, social, and economic behaviors: a panel study of twins and families. Twin Res Hum Genet 2015; 18: 243–255.
Vellante M, Baron-Cohen S, Melis M, Marrone M, Petretto DR, Masala C et al. The ‘Reading the Mind in the Eyes’ test: systematic review of psychometric properties and a validation study in Italy. Cogn Neuropsychiatry 2013; 18: 326–354.
Fernández-Abascal EG, Cabello R, Fernández-Berrocal P, Baron-Cohen S . Test–retest reliability of the ‘Reading the Mind in the Eyes’ test: a one-year follow-up study. Mol Autism 2013; 4: 33.
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR . Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–959.
Browning SR, Browning BL . Rapid and accurate haplotype phasing and missing-data inference for Whole-Genome Association Studies by use of localized haplotype clustering. Am J Hum Genet 2007; 81: 1084–1097.
Willer CJ, Li Y, Abecasis GR . METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N et al. Common genetic variants influence human subcortical brain structures. Nature 2015; 520: 224–229.
Bigdeli TB, Lee D, Webb BT, Riley BP, Vladimirov VI, Fanous AH et al. A simple yet accurate correction for winner’s curse can predict signals discovered in much larger genome scans. Bioinformatics 2016; 32: 2598–2603.
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–295.
Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T et al. OpenMx: an open source extended structural equation modeling framework. Psychometrika 2011; 76: 306–317.
Barbeira A, Shah KP, Torres JM, Wheeler HE, Torstenson ES, Edwards T et al MetaXcan: summary statistics based gene-level association method infers accurate PrediXcan results. 2016, available at http://biorxiv.org/content/early/2016/03/23/045260 abstract (accessed on 12 April 2016).
de Moor MHM, Costa PT, Terracciano A, Krueger RF, de Geus EJC, Toshiko T et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry 2012; 17: 337–349.
Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 2013; 340: 1467–1471.
Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016; 533: 539–542.
Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion M-JA et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry 2014; 19: 253–258.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45: 580–585.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 2014; 94: 677–694.
Kirkland RA, Peterson E, Baker CA, Miller S, Pulos S . Meta-analysis reveals adult female superiority in ‘Reading the mind in the eyes test’. N Am J Psychol 2013; 15: 121–146.
Knafo A, Uzefovsky F . Variation in empathy: the interplay of genetic and environmental factors. In: Legerstee M, Haley DW, Bornstein MH (eds). The infant mind: Origins of the social Brain. The Guilford Press: New York, 2013, pp 97–121.
Baker CA, Peterson E, Pulos S, Kirkland RA . Eyes and IQ: a meta-analysis of the relationship between intelligence and ‘Reading the Mind in the Eyes’. Intelligence 2014; 44: 78–92.
Ibanez A, Huepe D, Gempp R, Gutiérrez V, Rivera-Rei A, Toledo MI . Empathy, sex and fluid intelligence as predictors of theory of mind. Pers Individ Dif 2013; 54: 616–621.
Charlton RA, Barrick TR, Markus HS, Morris RG . Theory of mind associations with other cognitive functions and brain imaging in normal aging. Psychol Aging 2009; 24: 338–348.
Buitelaar JK, van der Wees M, Swaab-Barneveld H, van der Gaag RJ . Verbal memory and Performance IQ predict theory of mind and emotion recognition ability in children with autistic spectrum disorders and in psychiatric control children. J Child Psychol Psychiatry 1999; 40: 869–881.
Baron-Cohen S, Baldwin DA, Crowson M . Do children with autism use the speaker’s direction of gaze strategy to crack the code of language? Child Dev 1997; 68: 48–57.
Magalhães E, Costa P, Costa MJ . Empathy of medical students and personality: evidence from the Five-Factor Model. Med Teach 2012; 34: 807–812.
Beadle JN, Paradiso S, Salerno A, McCormick LM . Alexithymia, emotional empathy, and self-regulation in anorexia nervosa. Ann Clin Psychiatry 2013; 25: 107–120.
Brewer R, Cook R, Cardi V, Treasure J, Bird G . Emotion recognition deficits in eating disorders are explained by co-occurring alexithymia. R Soc open Sci 2015; 2: 140382.
Dinsdale N, Mokkonen M, Crespi B, Abu-Akel A, Abu-Akel A, Bailey AL et al. The ‘extreme female brain’: increased cognitive empathy as a dimension of psychopathology. Evol Hum Behav 2016; 37: 323–336.
Campanella F, Shallice T, Ius T, Fabbro F, Skrap M, Adolphs R et al. Impact of brain tumour location on emotion and personality: a voxel-based lesion-symptom mapping study on mentalization processes. Brain 2014; 137: 2532–2545.
Kemp J, Berthel M-C, Dufour A, Després O, Henry A, Namer IJ et al. Caudate nucleus and social cognition: neuropsychological and SPECT evidence from a patient with focal caudate lesion. Cortex 2013; 49: 559–571.
Abu-Akel A, Shamay-Tsoory S . Neuroanatomical and neurochemical bases of theory of mind. Neuropsychologia 2011; 49: 2971–2984.
Tossell K, Andreae LC, Cudmore C, Lang E, Muthukrishnan U, Lumsden A et al. Lrrn1 is required for formation of the midbrain-hindbrain boundary and organiser through regulation of affinity differences between midbrain and hindbrain cells in chick. Dev Biol 2011; 352: 341–352.
Warrier V, Toro R, Chakrabarti B, Litterman N, Hinds D, Bourgeron T et al. Genome-wide analyses of empathy and systemizing: heritability and correlates with sex, education, and psychiatric risk. Cold Spring Harbor Labs J 2016; bioRxiv preprint.
Chapman E, Baron-Cohen S, Auyeung B, Knickmeyer R, Taylor K, Hackett G . Fetal testosterone and empathy: evidence from the empathy quotient (EQ) and the ‘reading the mind in the eyes’ test. Soc Neurosci 2006; 1: 135–148.
Acknowledgements
This study was funded by grants from the Templeton World Charity Foundation, Inc., the Medical Research Council, the Wellcome Trust, the Autism Research Trust, the Institut Pasteur, the CNRS and the University Paris Diderot. V.W. is funded by St John’s College, Cambridge, and Cambridge Commonwealth Trust. The research was carried out in association with the National Institute for Health Research (NIHR) Collaboration for Leadership in Applied Health Research and Care East of England at Cambridgeshire and Peterborough NHS Foundation Trust. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The research was supported by the National Human Genome Research Institute of the National Institutes of Health (grant number R44HG006981). The National Science Foundation (grant numbers 0729493 and 0721707) supported the research on the Brisbane Longitudinal Twin Study. F.U. was supported by the British Friends of Haifa University, the Israel Science Foundation (grant no. 449/14), the British Friends of Hebrew University and the Joseph Levy Charitable Foundation. T.B. was supported by the Institut Pasteur, the University Paris Diderot, CNRS, and the Bettencourt-Schueller Foundation. We acknowledge with gratitude the generous support of Drs Dennis and Mireille Gillings in strengthening the collaboration between SBC and TB, and between Cambridge University and the Institut Pasteur. This publication was made possible through the support of a grant from the Templeton World Charity Foundation, Inc. The opinions expressed in this publication are those of the authors, and do not necessarily reflect the views of Templeton World Charity Foundation, Inc. We thank the research participants and employees of 23andMe for making this work possible. We also thank the volunteers of the Brisbane Longitudinal Twin Study and the NIHR Cambridge BioResource. Finally, we thank Silviu-Alin Bacanu, Hilary Finucane, Brendan Bulik-Sullivan, Carrie Allison, Michael Lombardo and Richard Bethlehem for helpful discussions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
David A Hinds and Nadia Litterman are employees of 23andMe, Inc. The remaining authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Warrier, V., Grasby, K., Uzefovsky, F. et al. Genome-wide meta-analysis of cognitive empathy: heritability, and correlates with sex, neuropsychiatric conditions and cognition. Mol Psychiatry 23, 1402–1409 (2018). https://doi.org/10.1038/mp.2017.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2017.122
This article is cited by
-
From verbal to physical violence: the different severity perception of stalking behaviors
Current Psychology (2024)
-
Sex differences in social cognition among individuals with schizophrenia and in healthy control participants: a secondary analysis of published data
Archives of Women's Mental Health (2024)
-
Cognitive Twists: The Coevolution of Learning and Genes in Human Cognition
Review of Philosophy and Psychology (2024)
-
Heterogeneous factors influence social cognition across diverse settings in brain health and age-related diseases
Nature Mental Health (2024)
-
The Social Shapes Test as a Self-Administered, Online Measure of Social Intelligence: Two Studies with Typically Developing Adults and Adults with Autism Spectrum Disorder
Journal of Autism and Developmental Disorders (2023)