Abstract
Chromosome 16p13.11 microduplication is a risk factor associated with various neurodevelopmental disorders such as attention-deficit/hyperactivity disorder, intellectual disabilities, developmental delay and autistic spectrum disorder. The underlying molecular mechanism of this genetic variation remained unknown, but its core genetic locus—conserved across mice and humans—contains seven genes. Here, we generated bacterial artificial chromosome-transgenic mice carrying a human 16p13.11 locus, and these mice showed the behavioral hyperactivity phenotype. We identified miR-484 as the responsible gene using a combination of expression and functional analyses. Mature miR-484 was expressed during active cortical neurogenesis, and overexpression of miR-484 decreased proliferation and increased neural progenitor differentiation in vivo. Luciferase screening identified the 3'-untranslated region of protocadherin-19 (Pcdh19) as a target of miR-484. The effect of miR-484 on neurogenesis was rescued by ectopic PCDH19 expression. These results demonstrate that miR-484 promotes neurogenesis by inhibiting PCDH19. Dysregulation of neurogenesis by imbalanced miR-484/PCDH19 expression contributes to the pathogenesis of 16p13.11 microduplication syndrome.
Similar content being viewed by others
Introduction
Several behavioral phenotypes are associated with 16p13.11 microduplication syndrome,1 including attention-deficit/hyperactivity disorder,2 intellectual disabilities,3 developmental delay4 and autistic spectrum disorder (ASD).5 The molecular mechanisms underlying the behavioral and pathological phenotypes of the microduplication syndrome remained unknown. Further, evidence for a common pathological endophenotype among patients is lacking. Previous studies provided evidence of a strong association between neurodevelopmental disorders and the 16p13.11 locus,6 although the responsible genes within the locus have not been identified. Intriguingly, both deletion and duplication of this locus appear to result in neuropsychological symptoms,7 suggesting that the locus has dosage-sensitive gene(s), which may have critical roles in brain development. These observations prompted us to explore the responsible genes in this locus. Advantageously, the locus is short enough to analyze the functions of all of its genes, although the genes for major synaptic proteins are not included.
Cortical neurogenesis is the primary and fundamental process underlying vertebrate brain development. Accumulating evidence suggests that abnormal neurogenesis causes abnormal behavioral phenotypes. Mice treated with the anti-mitotic agent methylazoxymethanol at E14.5 exhibit hyperactivity and cognitive dysfunction.8 In addition, animals lacking neurogenesis-related genes, like Tbr2 or Pax6, show behavioral abnormalities including hyperactivity9 or an autistic phenotype,10 respectively. More recently, a clear association between abnormal neurogenesis and synaptic disorders has been demonstrated. Overproduction of upper-layer neurons induces an autistic behavioral phenotype accompanied by an imbalance of excitatory and inhibitory synaptic connections in the cortex.11 Furthermore, research performed using an animal model of 21-trisomy showed that the pharmacological restoration of embryonic cortical neurogenesis rescued synaptic development.12 In the ASD brain, dysregulation of neuronal layer formation and differentiation occur presumably during the prenatal stage.13 Moreover, genetic risk factors in ASD include differentiation- and migration-related genes such as TBR1 and RELN.14 These findings support the notion that dysregulation of neurogenesis is one of the critical causes of neurodevelopmental disorders.
Based on the above-mentioned findings, we hypothesized that non-synaptic and dosage-sensitive candidate gene(s) are located in the 16p13.11 locus and that these genes control embryonic cortical neurogenesis or radial migration. The present study revealed the dosage-sensitive gene miR-484, which is located within the 16p13.11 common core locus. Insertion of the human 16p13.11 core locus into the mouse genome causes behavioral abnormalities and miR-484 upregulation. Moreover, we found that miR-484 controls cortical neurogenesis by inhibiting protocadherin-19 (PCDH19) translation and mRNA stabilization.
Materials and methods
Alignment of the duplication locus of 16p13.11 patients
We performed a database search in the DECIPHER database (https://decipher.sanger.ac.uk) and identified 162 patients with 16p13.11 microduplication syndrome. We excluded patients with multiple copy number variation loci or those without a phenotype in the brain. Finally, we obtained the duplication loci of 43 patients. We also examined 16p13.11 microduplication in the International Standards for Cytogenomic Arrays/ClinGen database (www.clinicalgenome.org/). Among the 105 patients, we included only those patients who had microduplication that was certified as ‘pathogenic’ or ‘likely pathogenic.’ Of these, we included 10 patients who exhibited the phenotype in the brain. In addition, we consulted two papers showing that 16p13.11 microduplication is important for neurodevelopmental disorders like attention-deficit/hyperactivity disorder.2, 15 After obtaining all duplication loci, we aligned them and manually determined the duplication core locus. All patients’ information is described in Supplementary Table 1.
Animals
All mice used in the in utero electroporation or in situ hybridization experiments were the slc-ICR strain (purchased from SLC Japan, Fukuoka, Japan). The mice were bred and maintained at the experimental animal facility of Osaka University Graduate School of Medicine and were killed with an overdose of a mixture of Vetorphale (0.5 mg ml−1, Meiji Seika Pharma, Tokyo, Japan), Dormicum (0.4 mg ml−1, Roche, Basel, Switzerland) and Domitor (0.03 mg ml−1, Orion, Espoo, Finland) by peritoneal injection. All procedures complied with the Osaka University Medical School Guidelines for the Care and Use of Laboratory Animals.
Generation of BAC-Tg mice
To generate bacterial artificial chromosome-transgenic (BAC-Tg) mice, we used a BAC clone (RP11-121O8) carrying a segment of human chromosome 16p13.11 that contained KIAA0430, MIR484 and NDE1. The BAC clone was injected into the BDF1 (C57BL6N/DBA-2 hybrid F1) embryos as previously described.16 The primers used for genotyping were designed against human-specific sequences (forward: 5′-AGCTCCTATAATCCCCGTGTGTTGTGG-3′, reverse: 5′-CTTACTGTGCCAGGAAAGCA GACCCTA-3′).
Open-field test
The open-field test apparatus was placed in a small soundproof box. Two-month-old mice were blindly and randomly obtained from their cages and individually placed in the center of the open-field test apparatus (50 × 50 × 40 cm, O’HARA & CO, Tokyo, Japan). For each box, a CCD camera was attached to the ceiling of each box. The distance traveled (cm) and time spent in the center (nine small squares in the center of 25 small squares) were recorded automatically using Image OF software (O’Hara). Data were collected for 10 min.
MicroRNA-related oligos
Negative control precursor (NCP; mirVana miRNA mimic, Negative control #1), miR-484 precursor (miR-484P; mirVana miRNA mimic), control inhibitor oligo (NCI; mirVana miRNA inhibitor, Negative control #1) and miR-484- specific inhibitor oligo (miR-484I), were all purchased from Thermo Fisher Scientific, Waltham, MA, USA.
Plasmids
See Supplementary Information.
In situ hybridization
Digoxigenin-labeled riboprobes were prepared by in vitro transcription, or miRCURY LNA microRNA Detection Probes for mature miR-484 or scrambled sequence were purchased from Exiqon (Vedbaek, Denmark). In situ hybridization was performed as described previously.17 In brief, the brains were removed, fixed overnight in 4% paraformaldehyde (PFA) in 0.1 m phosphate buffer, and one more overnight again in 30% sucrose/4% PFA in 0.1 m phosphate buffer and sectioned (at 40 μm) by a sliding microtome (Leica Microsystems, Wetzlar, Germany). Sections were hybridized with digoxigenin-labeled probe at 70 °C overnight. Excess probes were washed out and signals were detected with alkaline phosphatase-coupled antibody to digoxigenin (Roche) with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate as color reaction substrates. Some sections were processed further for immunohistochemistry.
For miR-484, the protocol was slightly modified. The brains were removed, freshly frozen in dry ice and sectioned at 20 μm with a cryostat (Thermo Fisher Scientific). Sections were hybridized with a digoxigenin-labeled probe at 50 °C overnight. The rest of the remaining procedures were the same as above.
Quantitative real-time PCR analysis
Tissues and cortical neurons were homogenized in Trizol (Thermo Fisher Scientific) and total RNA was purified using the RNeasy Micro Kit (Qiagen, Hilden, Germany). The microRNA-enriched fraction was purified by the miRVana miRNA Isolation kit (Thermo Fisher Scientific). The complementary DNA synthesis was performed with the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative Taqman real-time PCR analysis was performed with Taqman Universal Master Mix II (Thermo Fisher Scientific) and Taqman MicroRNA assays for miR-484 and sno-202 (control) or Taqman Gene expression assays for Gapdh or Pcdh19 (Thermo Fisher Scientific).
Cortical progenitor culture and transfection
The E12.5 cortical progenitors from ICR mice were cultured in the Neurobasal media (Thermo Fisher Scientific) containing 40 ng ml−1 fibroblast growth factor 2 (Promega, Fitchburgm, WI, USA), 2% B27 (Life Technologies), 120 mg ml−1 penicillin, 200 mg ml−1 streptomycin sulfate and 600 mg ml−1 glutamine, as described previously,18 at a density of 350 000 cells per well in four-well chamber slides. For transfection, 30 min after plating, a mix of 1 μg of reporter plasmid, 1.5 μl FugeneHD (Promega) and miRNA-related oligos at various final concentrations (10, 25, 100 nM) was added to cells in 100 μl of culture media.18
Cortical neuron culture and nucleofection
E14.5 or E15.5 mouse cortices were digested in 0.25% trypsin (Thermo Fisher Scientific) with DNase (1:1000, Sigma, Merck, Darmstadt, Germany) for 20 min at 37 °C, followed by dissociation in DMEM (Dulbecco’s Modified Eagle Medium) containing 10% fetal bovine serum, as described previously.19 The cells were washed and resuspended in 100 μl of Mouse Neuron Nucleofector Solution (Lonza, Basel, Switzerland) with 5 μl of 25 μM NCP, miR484P, NCI or miR484I. Immediately after electroporation, the cells were mixed with 500 μl of DMEM/F-12 containing 10% fetal bovine serum, and the cell suspension was transferred onto poly-l-lysine-coated dishes. The medium was exchanged with DMEM/F-12 (Thermo Fisher Scientific) containing 2% B27 supplement (Thermo Fisher Scientific) 12 h later.
Immunocytochemistry
Cortical progenitors were cultured for 2 days to assess proliferation and for 3 days to analyze in vitro differentiation. The cells were fixed for 10 min in 4% PFA in 0.1 m phosphate buffer, permeabilized with 0.2% NP-40 for 10 min and blocked in 6% goat serum and 0.5% bovine serum albumin in phosphate-buffered saline (−) for 1 h. The slides were incubated with primary antibodies overnight at 4 °C, followed by 1-h incubation with secondary antibodies at room temperature. Mouse anti-Ki-67 (1:200, BD Bioscience Franklin Lakes, NJ, USA), chick anti-green fluorescent protein (GFP) (1:400, Thermo Fisher Scientific), rabbit anti-GFP (1:400, Thermo Fisher Scientific), mouse Tuj1 (1:1000, Covance, Princeton, NJ, USA) and rabbit Tuj1 (1:1000, Covance) were used as primary antibodies. Alexa 488 goat anti-rabbit and anti-mouse immunoglobulin G (1:500, Thermo Fisher Scientific), and Alexa 568 anti-rabbit, and anti-mouse IgG immunoglobulin G (1:500, Thermo Fisher Scientific) were used as secondary antibodies. Finally, we counterstained with 4',6-diamidino-2-phenylindole (DAPI; (1:1000, Dojindo, Kumamoto, Japan) and mounted the slides using Fluorescence Mounting Medium (Agilent Technologies, Santa Clara, CA, USA). Images were acquired using a BX51 fluorescence microscope (Olympus, Tokyo, Japan).
In utero electroporation
In utero electroporation was performed on E13.5 or 14.5 embryos using a square-wave electroporator CUY21SC (NEPAGENE, Ichikawa, Japan) that delivered five 50-ms pulses of 30 or 35 V at 950-ms intervals, as described elsewhere.18 We injected a 2-μl mixture of 3 μg μl−1 nuclear EGFP expression vector with 50 μM NCP, miR-484P, NCI or miR-484I at a ratio of 1:1. For observing radial migration, 1 μg of pCAG-IRES-GFP, or pCAG-Nde1, or Fopnl-IRES-GFP expression vector were injected into the E14.5 embryo. For Pcdh19 shRNA experiments, 6.0 μg of the plasmids were injected that and consisted of a mixture of nuclear EGFP and Pcdh19 shRNA expression vectors at a ratio of 1:4. Trypan blue (Thermo Fisher Scientific) (0.05%) was co-injected as a tracer. For the differentiation assay, brains were dissected 2 days after electroporation. For the migration assays, brains were dissected at P0, 5 days after electroporation.
In vivo BrdU labeling
BrdU (Sigma) was administered intraperitoneally to pregnant mom mice at 50 mg kg−1 at E13.5. Forty-eight hours after injection, embryonic brains were dissected and then immersed overnight in 4% PFA. Tails were used for genotyping and gender identification by amplification of SRY gene.
Immunohistochemistry
Post-fixed E10.5, E12.5, E15.5, E16.5 and P0 mouse brains were sectioned at 20 μm. Sections were fixed in 4% PFA in 0.1 m PB for 15 min, followed by blocking and permeabilization with 10% bovine serum albumin and 0.3% Triton X-100, and then finally treated with the M.O.M. blocking kit according to the manufacturer’s instructions (Vector Laboratories, Burlingame, CA, USA). Mouse anti-Ki-67 (1:200, BD Bioscience), chick anti-GFP (1:400, Thermo Fisher Scientific), rabbit anti-GFP (1:400, Thermo Fisher Scientific), mouse (anti-phospho-histone H3 (pHH3)) (1:1000, Abcam, Cambridge, UK), rabbit anti-Pax6 (1:500, Merck Millipore, Billerica, MA, USA), rabbit anti-Tbr2 (1:500, Abcam), rabbit anti-PCDH19 (1:100, Merck), and mouse anti-SatB2 (1:200, Abcam) and anti-BrdU (1:200, Accurate Chemical and Scientific, Westbury, NY, USA) were used as primary antibodies. DAPI (1:1000, Dojindo) was used to counterstain the nuclei. For the proliferation and differentiation assays, sections with a similar anatomical EGFP distribution of EGFP expression were chosen selected for analysis, and a total of 3–5 sections were analyzed per embryo on a BX51 fluorescence microscope (Olympus) or FV-1200 laser-scanning confocal microscope (Olympus). The subventricular zone (SVZ) area was determined by DAPI staining, as described previously.19
Luciferase assay screening
Cells (2.5 × 104 HEK-293T, obtained from RIKEN BRC cell bank) were seeded in 24-well culture dishes and cultured for 1 day. Then, cells were transfected with 2 μl of Lipofectamine2000 (Thermo Fisher Scientific) and 20 ng of pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) in combination with 5 μl of 25 μM NCP or miR-484P. After 3 days, cells were lysed in lysis buffer of the Dual-Luciferase Reporter Assay System (Promega). The luciferase assay with triplicate samples was performed according to the manufacturer’s instructions with a Microplate Luminometer (CentroXS3 LB960, Berthold Technologies, Bad Wildbad, Germany). For the luciferase assay of the neurons, just after trituration of the cortices, 5 × 106 cells were nucleofected with 2 μg of pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) with 5 μl of 25 μM NCP or miR-484P, according to the manufacturer’s instructions (Lonza). Three days after nucleofection, neurons were analyzed as described above.
Western blotting
Transfected HEK-293T cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) containing protease inhibitors (Roche). Lysates were separated on 8% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene fluoride membranes. Membranes were blocked with 5% skim milk in phosphate-buffered saline-T (0.05% Tween-20) buffer for 60 min, and incubated with primary antibody at 4 °C overnight. The primary antibodies used for immunoblotting were rabbit anti-Myc (1:1000, Merck Millipore) and rabbit anti-PCDH19 (1:500, Merck). After washing with phosphate-buffered saline-T, membranes were incubated for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibodies (1:2000, Cell Signaling Technologies, Danvers, MA, USA). Signals were detected using enhanced chemiluminescence (ECL plus, GE Healthcare, Chicago, IL, USA), and images were obtained using a LAS-3000 image analyzer (Fuji Film, Tokyo, Japan).
Statistics
We chose the sample size according to the previous reports.17, 18, 19 Quantitative data are expressed as the mean±s.e.m. Biological replicates were used for each experiment. GraphPad Prism6 software (GraphPad Software) was used for all statistical analyses. Differences across all groups were analyzed with one-way analyses of variance (ANOVAs) followed by Tukey–Kramer tests. Differences between pairs of experimental groups pairs were analyzed with Student’s t-tests (two-tailed). P<0.05 was considered significant in all the experiments.
Results
Expression of the candidate genes responsible for 16p13.11 microduplication syndrome
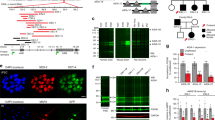
We first used an in silico approach to examine the candidate genes for 16p13.11 microduplication syndrome. As illustrated in Figure 1a and Supplementary Table 1, we identified a core gene locus that was shared among the neurodevelopmental syndromes. This locus was from 15551062 to 16194575 in human genome reference sequences GRCh37. These sequences contain seven genes (c16orf45, KIAA0430, MIR484, NDE1, MYH11, FOPNL (FGFR1OP N-terminal like), ABCC1). This gene locus is highly conserved between humans and mice (Figure 1b). We then analyzed the gene expression by in situ hybridization in embryonic and postnatal mouse brains to highlight the candidate genes. We found that five genes, including Minp (human C16ORF45),19 Marf1 (human KIAA0430), Mir484 (human MIR484), Fopnl (human FOPNL) and Nde1 (human NDE1), were expressed in the embryonic cortex (Figures 1c and 2j); additionally, Marf1 and Fopnl were expressed in the postnatal cortex. On the other hand, Myh11 and Abcc1 expression in the developing brain was below the detectable level. Therefore, we excluded Myh11 and Abcc1 from the list of candidate genes.
Candidate genes for 16p13.11 microduplication syndrome identified through expression and functional screening. (a) Each blue bar represents an individual duplication locus obtained from the DECIPHER/ISCA databases and previously reported papers.2, 15 The core locus is in between 15551062~16194575 and is flanked by two vertical black lines. (b) Comparison between Homo sapiens 16p13.11 locus and Mus musculus 16p13.11 locus. Blue bar represents the aligned core locus. Black bar shows each gene locus in the core region within the GRCh37 and GRCm38.p2 sequences. Minp, Marf1, MiR484, Nde1, Myh11, Fopnl and Abcc1 are located in the core locus. (c) Left-side images show sections of the rostral, middle and caudal parts of E13.5 brains and right-side images show P7 brains. Dashed squares represent high-magnification images of the dashed box in rostral sections of P7 brains. (d, f) Immunostaining for GFP (green) in combination with RG marker Pax6 (red) at SVZ (d), and BP marker Tbr2 (red) at SVZ (f) on E15.5, 2 days after electroporation of control (cagig) vectors or expressing vectors for Nde1 or Fopnl. (e, g) Quantification of the proportion of GFP-labeled cells positive for Pax6 (e) and Tbr2 (g). Differences are not statistically significant. (n=3, one-way ANOVA followed by Tukey–Kramer test). Scale bar: 50 μm. (h) Immunostaining for GFP (green) with DAPI on postnatal day 0, 5 days after electroporation. Cortical layers are determined by DAPI staining. WM, white matter; MZ, marginal zone.
miR-484 is the responsible gene for 16p13.11 microduplication syndrome. (a) Generation of the BAC-Tg mouse carrying three candidate genes: human KIAA0430, MIR-484 and NDE1. (b) Representative automatic traces of open-field exploration during a 10-min trial. WT: wild-type littermate, Tg: transgenic littermate, ♂: male, ♀: female. (c) Quantification of the total distance traveled during the trial. *P<0.05 (n=7 for transgenic male, n=13 for the others, one-way ANOVA followed by Tukey–Kramer test). (d) Mean percentage of time spent in the center of the open-field. (e) Relative expression of mature miR-484 in Tg (white) and WT (black) female E16.5 cortex. *P<0.05 (n=3, Student’s t-test). (f, g) Relative expression of mouse Marf1 (f) and Nde1 (g) in Tg (white) and WT (black) female E16.5 cortex. (h) Time course changes of mature miR-484. *P<0.05, ***P<0.001, ****P<0.0001 vs E10.5 sample (n=3, one-way ANOVA followed by Tukey–Kramer test). (i) High-magnification micrographs of in situ hybridization analysis of mature miR-484 (blue) on sagittal sections of E14.5 cortex (scale bar: 300 μm). (j) In situ hybridization analysis of mature miR-484 (blue) on sagittal sections of E14.5 whole embryos (scale bar: 1000 μm). SVZ, subventricular zone; GE, ganglionic eminence; SC, spinal cord; L, liver; T, thyroid; K, kidney; FD, finger digits; V, primordium of the follicle of vibrissa. (k) Experimental schedule for E12.5 cortical progenitor differentiation. 1 DIV: cells were harvested and cultured with FGF2 for 1 day. After 4 DIV: cells were harvested and cultured with FGF2 for 1 day followed by 3 days in culture without FGF2. (l) Relative expression of mature miR-484 in cortical progenitors cultured for 1 DIV (black) and for 4 DIV (white). **P<0.01 vs 1 DIV (n=3, one-way ANOVA followed by Tukey–Kramer test). (m) Relative expression of mature miR-484 in ES-derived glutamatergic progenitors (black) and neurons (white). **P<0.01 vs progenitors. (n=3, Student’s t-test).
Exclusion of the candidate genes NDE1 and FOPNL by functional analysis
To narrow down the list of candidate genes, we conducted gain-of-function experiments by overexpressing candidate wild-type genes in embryonic brains using in utero electroporation (Figure 1d). We adopted gain-of-function experiments as a screening method, as microduplication may lead to overexpression of the candidate gene.20, 21 NDE1 regulates progenitor proliferation through centrosome duplication and mitotic spindle assembly and is involved in neuronal radial migration,22 whereas FOPNL controls ciliogenesis.23 These findings suggest that NDE1 and FOPNL may have primary roles in progenitor proliferation, neurogenesis and migration. To confirm this, we performed gain-of-function experiments by overexpressing NDE1 and FOPNL. However, neither NDE1 nor FOPNL overexpression altered the number of Pax6-positive radial glia (RG) or Tbr2-positive basal progenitors (BPs) at the SVZ of the embryonic cortices (Figure 1d–g). Moreover, neither NDE1 nor FOPNL overexpression affected radial migration (Figure 1h). As Nde1 expression was restricted to the SVZ during the embryonic and postnatal periods (Figure 1c), we excluded the possibility that Nde1 regulates synaptogenesis or synaptic maintenance in neurons. These results exclude NDE1 and FOPNL from the list of candidate genes that presumably regulate early brain development including progenitor proliferation, neurogenesis and radial migration. Furthermore, we found that 2900011O08Rik (MINP) overexpression revealed a subtle phenotype in embryonic cortical development.19
Female BAC-Tg mice show hyperactivity and miR-484 overexpression
To test whether the duplication of this candidate locus causes the behavioral phenotype observed in individuals with 16p13.11 microduplication syndrome, we utilized the BAC transgenic approach.24 We established a BAC-Tg mouse carrying RP11-121O8 human BAC-DNA containing KIAA0430, MIR484 and NDE1 (Figure 2a). After confirming the insertion of the desired BAC-DNA locus by PCR (data not shown), we analyzed the general activity of the BAC-Tg mice using an open-field test. Female BAC-Tg mice showed substantially higher spontaneous locomotor activity compared with wild-type mice (Figure 2b and c), suggesting that the 16p13.11 microduplication containing the ‘three core genes’ leads to hyperactivity in female mice exposed to new environments.
To determine the responsible gene, we examined expression of the candidate genes. Expression of mature miR-484, but not Marf1 or Nde1, was upregulated in the E16.5 BAC-Tg mouse cortex (Figure 2e–g), hence we focused on miR-484 in the subsequent experiments.
Association of miR-484 expression with neurogenesis
Both in vitro and in vivo, miR-484 regulates multiple biological phenomena, including mitochondrial fission and cancer angiogenesis by targeting Fis125 and vascular endothelial growth factor B/vascular endothelial growth factor receptor 2,26 respectively. However, its role and expression during development remained undetermined. Thus, we performed quantitative real-time PCR analyses with cortical brain samples obtained during the developmental stages (E10.5, E12.5, E14.5, E16.5, P1, adult). Mature miR-484 and its primary transcript were highly expressed around E14.5 when neurogenesis was most active (Figure 2h). We also examined the miR-484 tissue distribution by in situ hybridization at E14.5 and detected miR-484 expression in the SVZ, ganglionic eminence, spinal cord, liver, thyroid, kidney, finger digits and primordium of the follicle of vibrissa (Figure 2i and j). A scrambled-control probe detected no signal in the same section (data not shown). These observations suggest that miR-484 is involved in the differentiation of neuronal progenitor cells but not in the synaptogenesis of mature neurons, as the critical period of synaptogenesis is around birth.27 To confirm this, we performed an in vitro differentiation assay of primary cortical progenitor cells and homogenous embryonic stem cell-derived glutamatergic progenitors.28 We dissociated cortical progenitors at E12.5 and cultured them for 1 day in vitro (DIV) with fibroblast growth factor 2 or 1 DIV with fibroblast growth factor 2 followed by 3 days without fibroblast growth factor 2 (Figure 2k). The expression of miR-484 at 4 DIV was 1.5 times higher than that at 1 DIV (Figure 2l). This difference was more obvious when we used embryonic stem cell-derived cortical progenitors for the differentiation assay (Figure 2m), presumably because they were homogenous populations with minimal neuronal contamination at the starting point.28 These results suggest that miR-484 is involved in neurogenesis.
miR-484 promotes in vitro differentiation of the cortical progenitors
To address whether miR-484 regulates differentiation of the cortical progenitors, we performed gain-of-function and loss-of-function experiments in dissociated cortical progenitors in vitro. We transfected E12.5 cortical progenitors with miR-484 precursor oligo (miR-484P) or miR-484 inhibitory oligo (miR-484I) to overexpress or knockdown miR-484 expression, respectively (Supplementary Figures 1a and b). Then, we performed immunostaining for the cell proliferation marker Ki-67 at 2 DIV and for the neuronal differentiation marker Tuj1 at 3 DIV18 (Figure 3a and c). Ectopic expression of miR-484P reduced Ki-67-positive cycling progenitors and increased Tuj1-positive cells in a dose-dependent manner (Figure 3b and d). Conversely, transfection with miR-484I increased Ki-67-positive cycling progenitors and decreased Tuj1-positive cells (Figure 3e–h). These results suggest that miR-484 positively regulates cortical progenitor differentiation in vitro.
miR-484 regulates neurogenesis in vitro and in vivo. (a) Immunocytochemical analysis for GFP (green), Ki-67 (red) and DAPI (blue) in E12.5 cortical progenitors 2 days after transfection of NCP (control) or miR484P with nuclear GFP (nGFP). (b) Quantification of the GFP-labeled Ki-67-positive cells. Data are represented as a percentage of the NCP sample value. *P<0.05, **P<0.01 (n=3, one-way ANOVA followed by Tukey–Kramer test). (c) Immunostaining for GFP, Tuj1 and DAPI at 3 days after transfection of NCP or miR484P with cytoplasmic GFP-expressing vector. (d) Quantification of the GFP-labeled Tuj1-positive cells. Data are represented as a percentage of the NCP sample value. *P<0.05, ***P<0.001 (n=3, one-way ANOVA followed by Tukey–Kramer test). (e) Immunostaining for GFP, Ki-67 and DAPI in E12.5 cortical progenitors 2 days after transfection of NCI (control) or miR484I with nGFP. (f) Quantification of the GFP-labeled Ki-67-positive cells (n=3, *P<0.05, **P<0.01, one-way ANOVA followed by Tukey–Kramer test). (g) Immunostaining for GFP, Tuj1 and DAPI at 3 days after transfection of NCI or miR484I with cytoplasmic GFP-expressing vector. (h) Quantification of the GFP-labeled Tuj1-positive cells (n=3, **P<0.01, one-way ANOVA followed by Tukey–Kramer test). (i, k) Quantification of the apoptotic GFP-labeled Tuj1-positive neurons transfected with NCP (control) or miR-484P (i), NCI or miR484I (k). Data are represented as a percentage of the control value (n=3, one-way ANOVA followed by Tukey–Kramer test). (j, l) Quantification of the apoptotic GFP-labeled Tuj1-negative progenitors transfected with NCP or miR484P (j), NCI or miR484I (l). Data are represented as a percentage of the control value (n=3, one-way ANOVA followed by Tukey–Kramer test).(m) Immunostaining for GFP (green), Ki-67 (red) and DAPI (blue) in SVZ at E16.5, 2 days after electroporation of NCP (control: white) or miR484P (black) together with nGFP-expressing vector at E14.5. Right images represent high-magnification views of the dashed box in the merged pictures. (n) Graph demonstrates the percentage of GFP-labeled cells positive for Ki-67 following overexpression of NCP (white) or miR484P (black). *P<0.05 vs NCP (n=3, Student’s t-test). Scale bar: 50 μm. (o, q, s, u) Immunostaining for GFP (green) with Ki-67 (red) at SVZ (o), mitotic marker pHH3 (red) at VZ (q), RG marker Pax6 (red) at SVZ (s) and BP marker Tbr2 (red) at SVZ (u) at E15.5, 2 days after electroporation of NCI or miR484I with GFP-expressing vector at E13.5. (p, r, t, v). Quantification of the proportion of GFP-labeled cells positive for Ki-67 (p), pHH3 (r), Pax6 (t) or Tbr2 (v). *P<0.05, **P<0.01, ***P<0.001 vs NCI (n=3 or 4, Student’s t-test). Scale bar: 50 μm.
To establish whether miR-484 affects progenitor survival, we examined the proportion of cell death by measuring apoptotic nuclei at 3 DIV after overexpressing miR-484P (Supplementary Figures 1c). After immunostaining with Tuj1 and DAPI, we quantified apoptotic nuclei in Tuj1-positive neurons and Tuj1-negative progenitors (Supplementary Figure 1c), as previously reported.29 However, ectopic miR-484P expression did not affect the survival of neurons or progenitors (Figure 3i and j) nor did miR-484I expression (Figure 3k and l). Therefore, we conclude that miR-484 regulates neurogenesis without affecting cell survival in vitro.
miR-484 controls in vivo BP production
Next, we assessed the function of miR-484 in vivo by performing in utero electroporation of a nuclear GFP-expressing vector with control or miR-484P into E14.5 cortices. Sections prepared at E16.5 were immunostained for Ki-67, GFP and DAPI (Figure 3m). We quantified the proportion of GFP-labeled Ki-67-positive proliferating cells at the ventricular zone (VZ) and SVZ (VZ/SVZ). The results showed that Ki-67-positive cycling progenitors were suppressed by miR-484P overexpression (Figure 3n). We then conducted loss-of-function experiments by in utero electroporation of miR-484I or control (NCI) with nuclear GFP-expressing plasmids into E13.5 cortices. In the E15.5 cortices, we examined the percentage of GFP-labeled Ki-67-positive cells at VZ/SVZ (Figure 3o), pHH3-positive cells at VZ (Figure 3q), Pax6-positive RG at VZ/SVZ (Figure 3s), and Tbr2-positive BPs at SVZ (Figure 3u). Suppression of the miR-484 inhibition increased Pax6-positive RG and pHH3-positive M-phase proliferating progenitors at VZ and increased Ki-67-positive cycling progenitors, but decreased differentiation into Tbr2-positive BPs (Figure 3p, r, t and v). These results show that miR-484 is required for in vivo differentiation into BPs.
miR-484 regulates translation and mRNA stability of Pcdh19
To delineate the mechanisms underlying the effects of miR-484, we performed target screening of miR-484 using the Targetscan software in silico, as previously described.30 Although miR-484 is not fully conserved from lower to higher animals, it is conserved among higher vertebrates like primates and rodents (Figure 4a). Therefore, targets were selected in an unbiased manner from 3′-untranslated region (UTR) sequences that were conserved at least between mice and humans. We selected 3′-UTR sequences from the top of ~20 genes and cloned each of them into a luciferase vector. After transfection of the luciferase vector with miR-484P or NCP into HEK-293T cells, luciferase activity was measured after 48 h in culture. The activity induced by the luciferase vector containing Pcdh19 3′-UTR was significantly depressed by miR-484P overexpression compared WITH the control (NCP) (Figure 4b). To confirm the specificity of the 3′-UTR sequence that was potentially targeted by miR-484, a target-seed sequence was mutated (Figure 4c). Mutation in the seed sequence completely abolished the miR-484P overexpression-induced luciferase activity depression (Figure 4d).
miR-484 inhibits both the translation and mRNA stability of Pcdh19. (a) miR-484 sequences in vertebrates are represented. The miR-484 sequence is conserved among higher vertebrates (yellow box). (b) Graph demonstrates the ratio of Firefly/Renilla luciferase activity in HEK-293T cells transfected with the indicated luciferase vector containing the 3′-UTR candidate sequence with NCP (black) or miR-484P (white). Data are represented as a fold of the control value. *P<0.05 (n=3, Student’s t-test). (c) Representation of wild-type (upper) and mismatch (lower) sequences of Pcdh19 3′-UTR. (d) The Firefly/Renilla luciferase activity ratio in HEK-293T cells overexpressed with empty, wild-type (Pcdh19) or mismatched sequence containing luciferase vector with NCP (black) or miR-484P (white). Data are represented as a fold of the NCP control value. *P<0.05 (n=3, Student’s t-test). (e) Firefly/Renilla luciferase activity ratio in neurons nucleofected with empty or wild-type (Pcdh19) sequence containing luciferase vector with NCP (black) or miR-484P (white). Data are represented as a fold of the control value. *P<0.05 (n=3, Student’s t-test). (f) Expression of Pcdh19 mRNA at 2 and 3 days after transfection with miR-484P or NCP in cortical neurons. ****P<0.0001 vs 2 DIV NCP (n=3, one-way ANOVA followed by Tukey–Kramer test). Data are represented as a fold of the NCP sample value at 2 DIV. (g) Time course changes of the expression of mature miR-484 and Pcdh19 mRNA. (h) Representative image showing in situ hybridization analysis of mature miR-484 (purple, white arrows) and PCDH19 protein (brown, black arrowheads or black arrows) on E12.5 cortical sections. Scale bar: 50 μm. (i) Relative expression of mouse Pcdh19 in transgenic (white) and WT (black) female E16.5 cortex. *P<0.05 (n=3, Student’s t-test). (j) Immunostaining for BrdU (green) with DAPI(blue) (k) Quantification of the proportion of BrdU-labeled cells (white arrows) at VZ+SVZ (left) and IZ+CP (right) ***P<0.001. Scale bar: 50 μm.
Next, we determined whether our observation held true in E15.5 cortical neurons. Neurons were transfected with luciferase vector and oligos by nucleofection and cultured for 2 days. The activity induced by the luciferase vector containing Pcdh19 3′-UTR was significantly depressed by miR-484P in the neurons (Figure 4e). Moreover, consistent with the notion that microRNAs suppress target translation and destabilize target mRNA,31 the amount of Pcdh19 mRNA was reduced by miR-484 overexpression in neurons at 2 and 3 DIV (Figure 4f).
To confirm these results in vivo, we analyzed the time course of Pcdh19 mRNA expression. We observed an intriguing reciprocal pattern of miR-484 and Pcdh19 mRNA expression (Figure 4g). When miR-484 was upregulated in embryonic cortices around E14.5 and 16.5, expression of Pcdh19 mRNA was downregulated. An opposite pattern of expression was observed at a later time point. Furthermore, the reciprocal miR-484 and PCDH19 protein expression was confirmed by in situ hybridization for miR-484 and immunohistochemistry for PCDH19 in E12.5 cortical sections (Figure 4h). M-phase progenitors at the ventricular surface expressed PCDH19 but not miR-484, and progenitors at the SVZ that were not in M-phase expressed miR-484 but not PCDH19. However, after differentiation from progenitors, PCDH19 expression recovered (Figure 4h). These findings demonstrate that miR-484 targets PCDH19 in vitro and in vivo. Consistent with this, female BAC-Tg mice showed reduced Pcdh19 mRNA expression in vivo (Figure 4i).
To determine whether abnormal neurogenesis would be observed in BAC-Tg mice, we administered BrdU to pregnant females at E13.5 and analyzed cortices of both genotypes 72 h later. We quantified the percentage of BrdU-labeled cells at VZ or SVZ and cells at intermediate zone or cortical plate (Figure 4j). We found a decreased percentage of BrdU-labeled RGs or BPs at VZ/SVZ (Figure 4k), but increased differentiated neurons at intermediate zone or cortical plate. Thus, BAC-Tg mice showed abnormal neurogenesis, consistent with our observations for miR-484 overexpression in vivo.
PCDH19 negatively regulates the differentiation of RG into BPs
PCDH19 is classified as a δ2-type protocadherin and is a member of the non-clustered-type protocadherin family.32 Many heterozygous mutations in PCDH19 cause EFMR (epilepsy and mental retardation limited to females),33 a congenital neurological disorder. PCDH19 is believed to have a role in neuronal migration in zebrafish. Although its postnatal expression pattern has been analyzed,34 its expression patterns and functions in the embryonic mammalian brain remain unknown. Therefore, we conducted immunohistochemical experiments to determine the expression of PCDH19 in E10.5 and 12.5 cortices (Figure 5a–c) by using a PCDH19-specific antibody (Supplementary Figure 2a). PCDH19-positive cells were pHH3-positive mitotic sox2-positive RG at the ventricular surface of E10.5 and E12.5 cortices (Figure 5a–c).
PCDH19 negatively regulates the differentiation of RG and rescues the miR-484 phenotype. (a) Immunostaining for PCDH19 (green), pHH3 (red) and DAPI (blue) in E10.5 cortex. (b) Magnified images of the boxed areas in (a) (white arrows: mitotic progenitors at VZ. white arrowheads: neurons at preplate). (c) Immunostaining for PCDH19 (green), Sox2 (red) and DAPI (blue) in E12.5 cortex (right image: magnified image of the boxed area in left image). (d, f, h, j) Immunostaining for GFP (green) with Ki-67 (red) at SVZ (d), mitotic marker pHH3 (red) at VZ (f), RG marker Pax6 (red) at SVZ (h) and BP marker Tbr2 (red) at SVZ (j) on E15.5, 2 days after electroporation of scrambled shRNA vector (Scr) or shRNA vector targeting Pcdh19 (PCDH19shRNA1) with nGFP-expressing vector at E13.5. (e, g, i, k) Quantification of the proportion of GFP-labeled cells positive for Ki-67 (e), pHH3 (g), Pax6 (i) or Tbr2 (k). *P<0.05, ***P<0.001 vs Scr (n=3 or 4, Student’s t-test). Scale bar: 50 μm. (l, n) Immunostaining for GFP (green) together with Pax6 (red) at SVZ (l), and Tbr2 (red) at SVZ (n) on E15.5, 2 days after electroporation of NCP or miR484P with control (cagig) vector or PCDH19-expressing vector (Pcdh19) at E13.5. (m, o) Quantification of the proportion of GFP-labeled cells positive for Pax6 (m) or Tbr2 (o). *P<0.05, **P<0.01, ***P<0.001 compared with NCP+cagig (n=3, one-way ANOVA followed by Tukey–Kramer test). Scale bar: 50 μm. (p) Schematic illustration showing that 16p13.11 microduplication causes dysregulation of miR-484/PCDH19 expression and abnormal neurogenesis, which may lead to hyperactivity.
These results encouraged us to investigate the role of PCDH19 in cell-cycle regulation and cortical progenitor differentiation. We developed a specific shRNA vector against Pcdh19 (PCDH19shRNA1) (Supplementary Figure 2b) and introduced PCDH19shRNA1 or scrambled-control vector (Scr) along with a nuclear GFP expression vector into the E13.5 cortex by in utero electroporation (Figure 5d–k). At E15.5, we quantified the proportion of GFP-labeled, Ki-67-positive cells at VZ/SVZ (Figure 5e), pHH3-positive cells at VZ (Figure 5g), Pax6-positive cells at VZ/SVZ (Figure 5i), and Tbr2-positive cells at SVZ (Figure 5k). PCDH19 inhibition decreased RG proliferation and increased the differentiation of RG into BPs. We conclude that PCDH19 suppresses the in vivo differentiation of RG into BPs.
miR-484 targets PCDH19 to regulate neurogenesis in vivo
The above results suggest that miR-484 controls neurogenesis through the negative regulation of Pcdh19 mRNA and protein expression. To establish the hierarchy of the signaling molecules, we performed in vivo rescue experiments. NCP with control plasmids or PCDH19-expression plasmids, or miR-484P with control plasmids or PCDH19-expression plasmids, were introduced into the E13.5 cortex by in utero electroporation. Two days after electroporation, embryos were analyzed by immunohistochemistry. Although miR-484P overexpression reduced GFP-labeled Pax6-positive cells and increased GFP-labeled Tbr2-positive cells (Figure 5l and n), this effect was completely abolished by PCDH19 overexpression without target 3′-UTR (Figure 5m and o). Collectively, these results imply that miR-484 positively regulates in vivo neurogenesis by inhibiting PCDH19.
Discussion
In this study, we first determined the large core region of the 16p13.11 microduplication locus using the in silico approach, and then established the candidate locus, which contained three genes. Duplication of this locus in female mice produced hyperactivity. Next, we identified the responsible gene as miR-484. miR-484 regulates embryonic cortical neurogenesis by targeting Pcdh19 mRNA and protein translation. These results imply that the pathogenesis of female 16p13.11 microduplication syndrome involves dysregulation of Pcdh19 expression.
The 16p13.11 microduplication syndrome is a recently described syndrome,1 and its molecular mechanism, candidate gene and pathogenesis remained largely unknown. NDE1 was suggested as a candidate gene for this syndrome,35 which is consistent with the finding that the centrosome deficiency induced by the disruption of Nde1 leads to cortical progenitor expansion failure and microcephaly.22 However, neither neurogenesis nor migration dysregulation was observed by overexpressing wild-type NDE1 using the in utero electroporation method. Moreover, the female-specific phenotype of the locus duplication cannot be explained by NDE1 dysregulation. These results indicate that NDE1 is not a candidate gene of 16p13.11 microduplication syndrome. However, in the case of microRNA, locus duplication may directly affect its gene dosage, as we and other groups have shown.20, 21
Based on our results, miR-484 likely targets only Pcdh19. Generally, microRNA has over a hundred potential targets, as predicted by software. However, the false-positive rate of prediction programs ranges from 24 to 70%.36 Therefore, experimental validation is necessary to determine the interactions of individual microRNA and any potential target mRNAs.36 Reporter assays and rescue experiments have been employed to demonstrate their direct links. Although we have shown the direct link between miR-484 and Pcdh19, it should be noted that other potential target genes of miR-484 might be involved in the abnormal neurogenesis.
Point mutation and deletion of PCDH19 affects female brains, but not male brains, except for the mosaic somatic mutation in male subjects.37 Currently, it remains unclear why the X-linked PCDH19 mutation affects mainly female brain development. As the mosaic mutation has been observed in male subjects, a cellular interference hypothesis has been proposed.37 As the upstream molecular mechanism of PCDH19, miR-484 expression might be critical in this process and may explain the mechanisms underlying this X-linked female disease, especially since we observed the hyperactivity phenotype in female transgenic animals.
It has been established that PCDH19 is a multi-functional adhesive protein that plays key roles in brain development at various stages. PCDH19 regulates neural progenitor proliferation, as we have shown, potentially acts as a synaptic adhesive protein, and may work as an axon collective and elongation factor, like PCDH17,38, 39 another δ2-type protocadherin. As each δ-type protocadherin is expressed in a distinct brain area,34 the loss-of-function of PCDH19 by a single mutation would not be compensated for by other protocadherins. Thus, changes in PCDH19 expression in the developing brain by mutations or 16p13.11 microduplication may cause abnormal formation of specific circuits.
Moreover, regarding the clinical aspects of patients with PCDH19-mutated EFMR, EFMR is often comorbid with attention-deficit/hyperactivity disorder and ASD.40, 41 Interestingly, PCDH19 mutations are also reported in male patients with ASD and intellectual disability.42 These clinical reports indicate that PCDH19 mutation causes not only epilepsy with intellectual disability but also various neurodevelopmental disorders. Growing evidence supports that PCDH19 is one of the genetic causes of neurodevelopmental disorders.
Finally, although each neurodevelopmental disorder is thought to be based on a different molecular mechanism that affects a distinct neuronal system, these disorders are often comorbid with each other. The most probable core neurobiological endophenotype that appears to be shared among these disorders is synaptic disruption, at least in autism14, 27 and intellectual disabilities.43, 44 However, this synaptic disruption might be the consequence of prior dysregulation of developmental phenomena, like neurogenesis. Although further investigation is necessary to show the interaction of abnormal neurogenesis with behavioral phenotypes, investigating the pathogenesis of 16p13.11 microduplication syndrome and determining the responsible genes will help better our understanding of the neurobiological endophenotype for other neurodevelopmental disorders.
References
Watson CT, Marques-Bonet T, Sharp AJ, Mefford HC . The genetics of microdeletion and microduplication syndromes: an update. Annu Rev Genomics Hum Genet 2014; 15: 215–244.
Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 2010; 376: 1401–1408.
Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat 2007; 28: 674–682.
Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 2014; 46: 1063–1071.
Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 2011; 70: 886–897.
Malhotra D, Sebat J . CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 2012; 148: 1223–1241.
Mefford HC, Batshaw ML, Hoffman EP . Genomics, intellectual disability, and autism. N Engl J Med 2012; 366: 733–743.
Berger-Sweeney J, Hohmann CF . Behavioral consequences of abnormal cortical development: insights into developmental disabilities. Behav Brain Res 1997; 86: 121–142.
Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev 2008; 22: 2479–2484.
Umeda T, Takashima N, Nakagawa R, Maekawa M, Ikegami S, Yoshikawa T et al. Evaluation of Pax6 mutant rat as a model for autism. PLoS One 2010; 5: e15500.
Fang WQ, Chen WW, Jiang L, Liu K, Yung WH, Fu AK et al. Overproduction of upper-layer neurons in the neocortex leads to autism-like features in mice. Cell Rep 2014; 9: 1635–1643.
Guidi S, Stagni F, Bianchi P, Ciani E, Giacomini A, De Franceschi M et al. Prenatal pharmacotherapy rescues brain development in a Down's syndrome mouse model. Brain 2014; 137: 380–401.
Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S et al. Patches of disorganization in the neocortex of children with autism. N Engl J Med 2014; 370: 1209–1219.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014; 515: 209–215.
Ramalingam A, Zhou XG, Fiedler SD, Brawner SJ, Joyce JM, Liu HY et al. 16p13.11 duplication is a risk factor for a wide spectrum of neuropsychiatric disorders. J Hum Genet 2011; 56: 541–544.
Nakanishi T, Ikawa M, Yamada S, Parvinen M, Baba T, Nishimune Y et al. Real-time observation of acrosomal dispersal from mouse sperm using GFP as a marker protein. FEBS Lett 1999; 449: 277–283.
Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci 2013; 16: 543–551.
Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A et al. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol 2010; 20: 2058–2065.
Zhang S, Kanemitsu Y, Fujitani M, Yamashita T . The newly identified migration inhibitory protein regulates the radial migration in the developing neocortex. Sci Rep 2014; 4: 5984.
Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell 2009; 137: 1235–1246.
Adamo A, Atashpaz S, Germain P, Zanella M, D'Agostino G, Albertin V et al. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat Genet 2015; 47: 132–141.
Feng Y, Walsh CA . Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 2004; 44: 279–293.
Sedjai F, Acquaviva C, Chevrier V, Chauvin JP, Coppin E, Aouane A et al. Control of ciliogenesis by FOR20, a novel centrosome and pericentriolar satellite protein. J Cell Sci 2010; 123: 2391–2401.
Han K, Holder JL Jr, Schaaf CP, Lu H, Chen H, Kang H et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature 2013; 503: 72–77.
Wang K, Long B, Jiao JQ, Wang JX, Liu JP, Li Q et al. miR-484 regulates mitochondrial network through targeting Fis1. Nat Commun 2012; 3: 781.
Vecchione A, Belletti B, Lovat F, Volinia S, Chiappetta G, Giglio S et al. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc Natl Acad Sci USA 2013; 110: 9845–9850.
Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM . Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 2011; 14: 285–293.
Fujiki R, Sato A, Hata K, Tashiro F, Yasuhara N, Miyazaki J et al. Improvement in protocol to generate homogeneous glutamatergic neurons from mouse embryonic stem cells reduced apoptosis. Biochem Biophys Res Commun 2013; 430: 604–609.
Dugani CB, Paquin A, Fujitani M, Kaplan DR, Miller FD . p63 antagonizes p53 to promote the survival of embryonic neural precursor cells. J Neurosci 2009; 29: 6710–6721.
Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci 2011; 14: 1125–1134.
Huntzinger E, Izaurralde E . Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011; 12: 99–110.
Kahr I, Vandepoele K, van Roy F . Delta-protocadherins in health and disease. Prog Mol Biol Transl Sci 2013; 116: 169–192.
Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet 2008; 40: 776–781.
Hertel N, Redies C . Absence of layer-specific cadherin expression profiles in the neocortex of the reeler mutant mouse. Cereb Cortex 2011; 21: 1105–1117.
Tropeano M, Andrieux J, Collier DA . Clinical utility gene card for: 16p13.11 microdeletion syndrome. Eur J Hum Genet 2014; 22. doi: 10.1038/ejhg.2013.230.
Thomson DW, Bracken CP, Goodall GJ . Experimental strategies for microRNA target identification. Nucleic Acids Res 2011; 39: 6845–6853.
Depienne C, Bouteiller D, Keren B, Cheuret E, Poirier K, Trouillard O et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet 2009; 5: e1000381.
Hayashi S, Inoue Y, Kiyonari H, Abe T, Misaki K, Moriguchi H et al. Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev Cell 2014; 30: 673–687.
Hoshina N, Tanimura A, Yamasaki M, Inoue T, Fukabori R, Kuroda T et al. Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron 2013; 78: 839–854.
Scheffer IE, Turner SJ, Dibbens LM, Bayly MA, Friend K, Hodgson B et al. Epilepsy and mental retardation limited to females: an under-recognized disorder. Brain 2008; 131: 918–927.
Higurashi N, Shi X, Yasumoto S, Oguni H, Sakauchi M, Itomi K et al. PCDH19 mutation in Japanese females with epilepsy. Epilepsy Res 2012; 99: 28–37.
Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 2009; 41: 535–543.
van Bokhoven H . Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet 2011; 45: 81–104.
Zoghbi HY, Bear MF . Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol 2012; 4; doi: 10.1101/cshperspect.a009886.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science (25221309) to TY and a Health Labor Sciences Research Grant and Takeda Science Foundation Research Grant to MF. We thank Dr Yukio Kawahara for the helpful advice regarding the RNA experiments. We thank NPO Biotechnology Research and Development for the technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Fujitani, M., Zhang, S., Fujiki, R. et al. A chromosome 16p13.11 microduplication causes hyperactivity through dysregulation of miR-484/protocadherin-19 signaling. Mol Psychiatry 22, 364–374 (2017). https://doi.org/10.1038/mp.2016.106
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2016.106
This article is cited by
-
Neuronal network activity and connectivity are impaired in a conditional knockout mouse model with PCDH19 mosaic expression
Molecular Psychiatry (2023)
-
16p13.11 microdeletion/microduplication in fetuses: investigation of associated ultrasound phenotypes, genetic anomalies, and pregnancy outcome follow-up
BMC Pregnancy and Childbirth (2022)
-
Implications of germline copy-number variations in psychiatric disorders: review of large-scale genetic studies
Journal of Human Genetics (2021)
-
High-resolution chromosomal microarray analysis for copy-number variations in high-functioning autism reveals large aberration typical for intellectual disability
Journal of Neural Transmission (2020)
-
Reversal of proliferation deficits caused by chromosome 16p13.11 microduplication through targeting NFκB signaling: an integrated study of patient-derived neuronal precursor cells, cerebral organoids and in vivo brain imaging
Molecular Psychiatry (2019)