Abstract
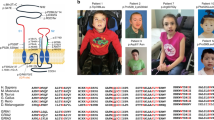
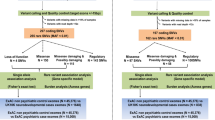
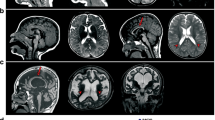
Phenotypic and genetic heterogeneity is predominant in autism spectrum disorders (ASD), for which the molecular and pathophysiological bases are still unclear. Significant comorbidity and genetic overlap between ASD and other neurodevelopmental disorders are also well established. However, little is understood regarding the frequent observation of a wide phenotypic spectrum associated with deleterious mutations affecting a single gene even within multiplex families. We performed a clinical, neurophysiological (in vivo electroencephalography—auditory-evoked related potentials) and genetic (whole-exome sequencing) follow-up analysis of two families with known deleterious NLGN4X gene mutations (either truncating or overexpressing) present in individuals with ASD and/or with intellectual disability (ID). Complete phenotypic evaluation of the pedigrees in the ASD individuals showed common specific autistic behavioural features and neurophysiological patterns (abnormal MisMatch Negativity in response to auditory change) that were absent in healthy parents as well as in family members with isolated ID. Whole-exome sequencing in ASD patients from each family identified a second rare inherited genetic variant, affecting either the GLRB or the ANK3 genes encoding NLGN4X interacting proteins expressed in inhibitory or in excitatory synapses, respectively. The GRLB and ANK3 mutations were absent in relatives with ID as well as in control databases. In summary, our findings provide evidence of a double-hit genetic model focused on excitatory/inhibitory synapses in ASD, that is not found in isolated ID, associated with an atypical in vivo neurophysiological pattern linked to predictive coding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders 5th (ed). American Psychiatric Press: Washington, DC, USA, 2013.
Berg JM, Geschwind DH . Autism genetics: searching for specificity and convergence. Genome Biol 2012; 13: 247.
Huguet G, Ey E, Bourgeron T . The genetic landscapes of autism spectrum disorders. Annu Rev Hum Genomics Hum Genet 2013; 14: 191–213.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014; 515: 209–215.
Betancur C . Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res 2011; 1380: 42–77.
Krumm N, O’Roak BJ, Shendure J, Eichler EE . A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci 2014; 37: 95–105.
Moraine C, Bonnet-Brilhault F, Laumonnier F, Gomot M . Could autism with mental retardation result from digenism and frequent de novo mutations? World J Biol Psychiatry 2009; 10: 1030–1036.
Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet 2010; 87: 316–324.
Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012; 485: 237–241.
Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012; 485: 242–245.
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012; 485: 246–250.
Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J et al. De novo gene disruptions in children on the autistic spectrum. Neuron 2012; 74: 285–289.
Wacongne C, Changeux JP, Dehaene S . A neuronal model of predictive coding accounting for the mismatch negativity. J Neurosci 2012; 32: 3665–3678.
Gomot M, Blanc R, Clery H, Roux S, Barthelemy C, Bruneau N . Candidate electrophysiological endophenotypes of hyper-reactivity to change in autism. J Autism Dev Disord 2011; 41: 705–714.
Gomot M, Giard MH, Adrien JL, Barthelemy C, Bruneau N . Hypersensitivity to acoustic change in children with autism: electrophysiological evidence of left frontal cortex dysfunctioning. Psychophysiology 2002; 39: 577–584.
Schroeder CE, Javitt DC, Steinschneider M, Mehta AD, Givre SJ, Vaughan HG Jr et al. N-methyl-D-aspartate-enhancement of phasic responses in primate neocortex. Exp Brain Res 1997; 114: 271–278.
Kreitschmann-Andermahr I, Rosburg T, Demme U, Gaser E, Nowak H, Sauer H . Effect of ketamine on the neuromagnetic mismatch field in healthy humans. Brain Res Cogn Brain Res 2001; 12: 109–116.
Korostenskaja M, Nikulin VV, Kicić D, Nikulina AV, Kähkönen S . Effects of NMDA receptor antagonist memantine on mismatch negativity. Brain Res Bull 2007; 72: 275–283.
Lahti AC, Weiler MA, Tamara Michaelidis BA, Parwani A, Tamminga CA . Effects of ketamine in normal and schizophrenic volunteers. Neuropsychopharmacology 2001; 25: 455–467.
Krystal JH, Bennett A, Abi-Saab D, Belger A, Karper LP, D'Souza DC et al. Dissociation of ketamine effects on rule acquisition and rule implementation: possible relevance to NMDA receptor contributions to executive cognitive functions. Biol Psychiatry 2000; 47: 137–143.
Gallinat J, Kunz D, Lang UE, Neu P, Kassim N, Kienast T et al. Association between cerebral glutamate and human behaviour: the sensation seeking personality trait. Neuroimage 2007; 34: 671–678.
Umbricht D, Koller R, Vollenweider FX, Schmid L . Mismatch negativity predicts psychotic experiences induced by NMDA receptor antagonist in healthy volunteers. Biol Psychiatry 2002; 51: 400–406.
Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM . Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 2004; 119: 1013–1026.
Hoon M, Soykan T, Falkenburger B, Hammer M, Patrizi A, Schmidt KF et al. Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proc Natl Acad Sci USA 2011; 108: 3053–3058.
Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet 2004; 74: 552–557.
Daoud H, Bonnet-Brilhault F, Védrine S, Demattéi MV, Vourc'h P, Bayou N et al. Autism and nonsyndromic mental retardation associated with a de novo mutation in the NLGN4X gene promoter causing an increased expression level. Biol Psychiatry 2009; 66: 906–910.
Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 2013; 493: 216–220.
Chung SK, Bode A, Cushion TD, Thomas RH, Hunt C, Wood SE et al. GLRB is the third major gene of effect in hyperekplexia. Hum Mol Genet 2013; 22: 927–940.
Thomas RH, Chung SK, Wood SE, Cushion TD, Drew CJ, Hammond CL et al. Genotype-phenotype correlations in hyperekplexia: apnoeas, learning difficulties and speech delay. Brain 2013; 136: 3085–3095.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP . Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012; 7: e46688.
Ng PC, Henikoff S . Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863–874.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Schwarz JM, Rödelsperger C, Schuelke M, Seelow D . MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576.
Yasunaga M, Ipsaro JJ, Mondragón A . Structurally similar but functionally diverse ZU5 domains in human erythrocyte ankyrin. J Mol Biol 2012; 417: 336–350.
Wang C, Yu C, Ye F, Wei Z, Zhang M . Structure of the ZU5-ZU5-UPA-DD tandem of ankyrin-B reveals interaction surfaces necessary for ankyrin function. Proc Natl Acad Sci USA 2012; 109: 4822–4827.
Bi C, Wu J, Jiang T, Liu Q, Cai W, Yu P et al. Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility. Hum Mutat 2012; 33: 1635–1638.
Iqbal Z, Vandeweyer G, van der Voet M, Waryah AM, Zahoor MY, Besseling JA et al. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet 2013; 22: 1960–1970.
Rasband MN . The axon initial segment and the maintenance of neuronal polarity. Nat Rev Neurosci 2010; 11: 552–562.
Smith KR, Kopeikina KJ, Fawcett-Patel JM, Leaderbrand K, Gao R, Schürmann B et al. Psychiatric Risk Factor ANK3/Ankyrin-G Nanodomains Regulate the Structure and Function of Glutamatergic Synapses. Neuron 2014; 84: 399–415.
Durak O, de Anda FC, Singh KK, Leussis MP, Petryshen TL, Sklar P et al. Ankyrin-G regulates neurogenesis and Wnt signaling by altering the subcellular localization of β-catenin. Mol Psychiatry 2014; 20: 388–397.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014; 515: 216–221.
Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry 2009; 66: 947–956.
Stessman HA, Bernier R, Eichler EE . Genotype-First Approach to Defining the Subtypes of a Complex Disease. Cell 2014; 156: 872–877.
Hoischen A, Krumm N, Eichler EE . Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci 2014; 17: 764–772.
Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 2014; 158: 263–276.
Laumonnier F, Nguyen LS, Jolly L, Raynaud M, Gecz J . UPF3B gene and nonsense-mediated mRNA decay in autism spectrum disorders Patel VB, Preedy VR, Martin CR (eds). Comprehensive Guide to Autism. Springer Science+Business Media: New York, USA, 2014; pp 1663–1678.
Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 2013; 155: 1008–1021.
Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M et al. Spatio-temporal transcriptome of the human brain. Nature 2011; 478: 483–489.
Miller JA, Ding S, Sunkin SM, Smith KA, Ng L, Szafer A et al. Transcriptional landscape of the prenatal human brain. Nature 2014; 508: 199–206.
Acknowledgements
We would like to thank the patients and their respective family for their collaboration in this project. We thank the 'PPF Analyses des systèmes biologiques' Department of the University of Tours for providing technical support for sequencing experimental procedures. This study received funding from INSERM, University François-Rabelais of Tours, EU FP7 GENCODYS project (to FL, Grant N°241995), Fondation de France (to FL and FBB, Grant N°2012-33662), and from Association for the Development of Neurogenetics (ADN). The BrainSpan project (Atlas of the Developing Human Brain, 2011) was funded by ARRA Awards 1RC2MH089921-01, 1RC2MH090047-01, and 1RC2MH089929-01.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
About this article
Cite this article
Bonnet-Brilhault, F., Alirol, S., Blanc, R. et al. GABA/Glutamate synaptic pathways targeted by integrative genomic and electrophysiological explorations distinguish autism from intellectual disability. Mol Psychiatry 21, 411–418 (2016). https://doi.org/10.1038/mp.2015.75
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2015.75
This article is cited by
-
Roles and mechanisms of ankyrin-G in neuropsychiatric disorders
Experimental & Molecular Medicine (2022)
-
22q13 deletion syndrome: communication disorder or autism? Evidence from a specific clinical and neurophysiological phenotype
Translational Psychiatry (2018)
-
Translational use of event-related potentials to assess circuit integrity in ASD
Nature Reviews Neurology (2017)