Abstract
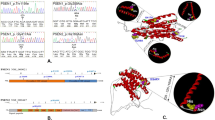
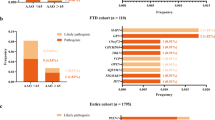
Performing exome sequencing in 14 autosomal dominant early-onset Alzheimer disease (ADEOAD) index cases without mutation on known genes (amyloid precursor protein (APP), presenilin1 (PSEN1) and presenilin2 (PSEN2)), we found that in five patients, the SORL1 gene harbored unknown nonsense (n=1) or missense (n=4) mutations. These mutations were not retrieved in 1500 controls of same ethnic origin. In a replication sample, including 15 ADEOAD cases, 2 unknown non-synonymous mutations (1 missense, 1 nonsense) were retrieved, thus yielding to a total of 7/29 unknown mutations in the combined sample. Using in silico predictions, we conclude that these seven private mutations are likely to have a pathogenic effect. SORL1 encodes the Sortilin-related receptor LR11/SorLA, a protein involved in the control of amyloid beta peptide production. Our results suggest that besides the involvement of the APP and PSEN genes, further genetic heterogeneity, involving another gene of the same pathway is present in ADEOAD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Campion D, Dumanchin C, Hannequin D, Dubois B, Belliard S, Puel M et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet 1999; 65: 664–670.
Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 2006; 38: 24–26.
Rovelet-Lecrux A, Legallic S, Wallon D, Flaman JM, Martinaud O, Bombois S et al. A genome wide study reveals rare CNVs exclusive to extreme phenotypes of Alzheimer disease. Eur J Hum Genet; advance online publication 14 December 2011 (e-pub ahead of print).
Alperovitch A, Amouyel P, Dartigues JF, Ducimetière P, Mazoyer B, Ritchie K et al. [Epidemiological studies on aging in France: from the PAQUID study to the Three-City study]. C R Biol 2002; 325: 665–672.
Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 2007; 39: 168–177.
Reitz C, Cheng R, Rogaeva E, Lee JH, Tokuhiro S, Zou F et al. Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch Neurol 2011; 68: 99–106.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D . MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576.
Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D et al. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry 2006; 45: 2618–2628.
Hermey G . The Vps10p-domain receptor family. Cell Mol Life Sci 2009; 66: 2677–2689.
Ng SB, Nickerson DA, Bamshad MJ, Shendure J . Massively parallel sequencing and rare disease. Hum Mol Genet 2010; 19: R119–R124.
Acknowledgements
This work was supported by a grant from the Clinical Research Hospital Program from the French Ministry of Health (GMAJ, PHRC 2008/067), to DH and DC, and sponsored by the University Hospital of Rouen. We thank the Integragen society, which performed exome sequencing. We thank the LITIS and the TIBS team for providing bioinformatics support, André Blavier for the Alamut software, Mario Tosi for helpful discussions, and Tracey Avequin for editing the manuscript.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Appendix A
Appendix A
PHRC GMAJ collaborators
The investigators of the French GMAJ project include Didier Hannequin, Dominique Campion, Olivier Martinaud, Lucie Guyant-Maréchal and David Wallon (Centre Hospitalo Universitaire (CHU), Rouen); Olivier Godefroy and Candice Picard (CHU Amiens); Frédérique Etcharry-Bouyx (CHU Angers); Eric Berger (CHU Besancon); Jean-Francois Dartigues and Sophie Auriacombe (CHU Bordeaux); Vincent de la Sayette (CHU Caen); Francois Sellal (CH Colmar); Olivier Rouaud and Christel Thauvin (CHU Dijon); Olivier Moreaud (CHU Grenoble); Stéphanie Bombois, Adeline Rollin-Sillaire, Marie-Anne Mackowiak and Florence Pasquier (CHU Lille); Isabelle Roullet-Solignac and Alain Vighetto (CHU Lyon); Mira Didic, Olivier Félician and Mathieu Ceccaldi (CHU Marseille); Audrey Gabelle and Jacques Touchon (CHU Montpellier); Martine Vercelletto and Claire Boutoleau-Bretonnière (CHU Nantes); Pierre Labauge and Giovanni Castelnovo (CHU Nimes); Claire Paquet and Jacques Hugon (CHU Lariboisière); Agnès Michon, Isabelle Le Ber and Bruno Dubois (CHU La Salpêtrière, Paris); Catherine Thomas-Antérion (CHU Saint-Etienne); Frédéric Blanc and Christine Tranchant (CHU Strasbourg); Jérémie Pariente, Michèle Puel and Jean-Francois Demonet (CHU Toulouse); Caroline Hommet and Karl Mondon (CHU Tours); Hélène Mollion and Bernard Croisile (CMRR CHU Lyon); Mathilde Sauvée (CHU Nancy); Gaelle Godenèche and Foucauld De Boisgueheneuc (CHU Poitiers).
Rights and permissions
About this article
Cite this article
Pottier, C., Hannequin, D., Coutant, S. et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry 17, 875–879 (2012). https://doi.org/10.1038/mp.2012.15
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2012.15
Keywords
This article is cited by
-
A familial missense variant in the Alzheimer’s disease gene SORL1 impairs its maturation and endosomal sorting
Acta Neuropathologica (2024)
-
Mutated Toll-like receptor 9 increases Alzheimer’s disease risk by compromising innate immunity protection
Molecular Psychiatry (2023)
-
Step by step: towards a better understanding of the genetic architecture of Alzheimer’s disease
Molecular Psychiatry (2023)
-
Genetic Phenotypes of Alzheimer’s Disease: Mechanisms and Potential Therapy
Phenomics (2023)
-
Mapping the genetic landscape of early-onset Alzheimer’s disease in a cohort of 36 families
Alzheimer's Research & Therapy (2022)