Abstract
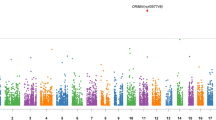
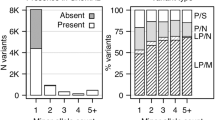
Large, rare copy number variants (CNVs) have been implicated in a variety of psychiatric disorders, but the role of CNVs in recurrent depression is unclear. We performed a genome-wide analysis of large, rare CNVs in 3106 cases of recurrent depression, 459 controls screened for lifetime-absence of psychiatric disorder and 5619 unscreened controls from phase 2 of the Wellcome Trust Case Control Consortium (WTCCC2). We compared the frequency of cases with CNVs against the frequency observed in each control group, analysing CNVs over the whole genome, genic, intergenic, intronic and exonic regions. We found that deletion CNVs were associated with recurrent depression, whereas duplications were not. The effect was significant when comparing cases with WTCCC2 controls (P=7.7 × 10−6, odds ratio (OR) =1.25 (95% confidence interval (CI) 1.13–1.37)) and to screened controls (P=5.6 × 10−4, OR=1.52 (95% CI 1.20–1.93). Further analysis showed that CNVs deleting protein coding regions were largely responsible for the association. Within an analysis of regions previously implicated in schizophrenia, we found an overall enrichment of CNVs in our cases when compared with screened controls (P=0.019). We observe an ordered increase of samples with deletion CNVs, with the lowest proportion seen in screened controls, the next highest in unscreened controls and the highest in cases. This may suggest that the absence of deletion CNVs, especially in genes, is associated with resilience to recurrent depression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Greenberg P, Stiglin L, Finkelstein S, Berndt E . The economic burden of depression in 1990. J Clin Psychiatry 1993; 54: 405–418.
Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA 2003; 289: 3095–3105.
Harris EC, Barraclough B . Excess mortality of mental disorder. Br J Psychiatry 1998; 173: 11–53.
Farmer A, Harris T, Redman K, Sadler S, Mahmood A, McGuffin P . Cardiff depression study. A sib-pair study of life events and familiality in major depression. Br J Psychiatry 2000; 176: 150–155.
McGuffin P, Katz R, Watkins S, Rutherford J . A hospital-based twin register of the heritability of DSM-IV unipolar depression. Arch Gen Psychiatry 1996; 53: 129–136.
Uher R . The role of genetic variation in the causation of mental illness: an evolution-informed framework. Mol Psychiatry 2009; 14: 1072–1082.
Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry 2011; 16: 516–532.
Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K et al. Genome-wide association study of major recurrent depression in the UK population. Am J Psychiatry 2010; 167: 949–957.
Feuk L, Carson AR, Scherer SW . Structural variation in the human genome. Nat Rev Genet 2006; 7: 85–97.
Cook EHJ, Scherer SW . Copy-number variations associated with neuropsychiatric conditions. Nature 2008; 455: 919–923.
Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S et al. Large recurrent microdeletions associated with schizophrenia. Nature 2008; 455: 232–236.
Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 2010; 376: 1401–1408.
International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008; 455: 237–241.
Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet 2009; 18: 1497–1503.
Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T et al. Strong association of de novo copy number mutations with autism. Science 2007; 316: 445–449.
Shinawi M, Schaaf CP, Bhatt SS, Xia Z, Patel A, Cheung SW et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet 2009; 41: 1269–1271.
Farmer A, Breen G, Brewster S, Craddock N, Gill M, Korszun A et al. The Depression Network (DeNT) Study: methodology and sociodemographic characteristics of the first 470 affected sibling pairs from a large multi-site linkage genetic study. BMC Psychiatry 2004; 4: 42.
Gaysina D, Cohen S, Craddock N, Farmer A, Hoda F, Korszun A et al. No association with the 5,10-methylenetetrahydrofolate reductase gene and major depressive disorder: results of the depression case control (DeCC) study and a meta-analysis. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 699–706.
Uher R, Maier W, Hauser J, Marusic A, Schmael C, Mors O et al. Differential efficacy of escitalopram and nortriptyline on dimensional measures of depression. Br J Psychiatry 2009; 194: 252–259.
Wing JK, Babor T, Brugha T, Burke J, Cooper JE, Giel R et al. SCAN schedules for clinical assessment in neuropsychiatry. Arch Gen Psychiatry 1990; 47: 589–593.
McGuffin P, Katz R, Aldrich J . Past and present state examination: the assessment of ‘lifetime ever’ psychopathology. Psychol Med 1986; 16: 461–465.
Beck AT, Steer RA . Internal consistencies of the original and revised Beck Depression Inventory. J Clin Psychol 1984; 40: 1365–1367.
Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007; 17: 1665–1674.
StataCorp. Stata Statistical Software: Release 10, 2007.
Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan P et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
Kirov G . The role of copy number variation in schizophrenia. Expert rev neurother 2010; 10: 25–32.
Glessner JT, Wang K, Sleiman PM, Zhang H, Kim CE, Flory JH et al. Duplication of the SLIT3 locus on 5q35.1 predisposes to major depressive disorder. PLoS One 2010; 5: e15463.
Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, Green EK et al. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch Gen Psychiatry 2010; 67: 318–327.
McQuillin A, Bass N, Anjorin A, Lawrence J, Kandaswamy R, Lydall G et al. Analysis of genetic deletions and duplications in the University College London bipolar disorder case control sample. Eur J Hum Genet 2011; 19: 588–592.
McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 2009; 41: 1223–1227.
Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 2008; 358: 667–675.
Robin NH, Shprintzen RJ . Defining the clinical spectrum of deletion 22q11.2. J Pediatr 2005; 147: 90–96.
Kirov G, Rujescu D, Ingason A, Collier DA, O’Donovan MC, Owen MJ . Neurexin 1 (NRXN1) deletions in schizophrenia. Schizophr bull 2009; 35: 851.
Rujescu D, Ingason A, Cichon S, Pietiläinen OP, Barnes MR, Toulopoulou T et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum mol genet 2009; 18: 988–996.
Gottesman II, Shields J . A polygenic theory of schizophrenia. Proc Natl Acad Sci USA 1967; 58: 199–205.
Plomin R, Haworth CM, Davis OS . Common disorders are quantitative traits. Nat Rev Genet 2009; 10: 872–878.
Acknowledgements
This study was funded by a joint grant from the UK Medical Research Council and GlaxoSmithKline (G0701420). James Rucker was supported by a fellowship from the Wellcome Trust (086635). Alexandra Schosser was supported by the Erwin-Schroedinger Fellowship (J2647) of the Austrian Science Funds. Alexandra Schosser, Inti Pedroso and Sarah Cohen-Woods received financial support from the National Institute for Health Research (NIHR) Specialist Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and the Institute of Psychiatry, King's College London. Margarita Rivera was supported by a Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme. The GENDEP study was funded by a European Commission Framework 6 grant, EC Contract Ref.: LSHB-CT-2003-503428 and GlaxoSmithKline contributed by funding an add-on project in the London centre. The population-based study in Lausanne was supported by three grants from the Swiss National Science Foundation (No. 3200B0-105993, No. 3200B0-118308, No. 33CSCO-122661) and from GlaxoSmithKline (Psychiatry Center of Excellence for Drug Discovery and Genetics Division, Drug Discovery Verona, R&D). Rudolf Uher and Peter McGuffin are supported by a grant from the European Commission (Grant Agreement No. 115008). Genotyping was performed at the Centre Nationale De Genotypage, Evry, Paris, with acknowledgement to Simon Heath, Ivo Gut and Mark Lathrop. We acknowledge the contribution of phase 2 of the Wellcome Trust Case Control Consortium in providing access to control data sets from the 1958 British birth cohort and the national blood service cohort.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The sponsors of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report; however, Pierandrea Muglia and Michael Barnes were employed by GSK when the research was performed. James Rucker, Gerome Breen and Peter McGuffin had full access to all data in the study and had final responsibility for the decision to submit for publication. Katherine Aitchison, Anne Farmer and Peter McGuffin have received consultancy fees and honoraria for participating in expert panels for pharmaceutical companies including GlaxoSmithKline. Anne Farmer has received travel and subsistence from GlaxoSmithKline to attend principal investigator planning, training and inter-rater reliability meetings. Katherine Aitchison also declares interests through Advisory Boards for Johnson & Johnson, Lundbeck, Roche Diagnostics, and Bristol-Myers Squibb; membership of Bristol-Myers Squibb UK Steering group 2003 to present; consultancy work for Roche Diagnostics, Johnson & Johnson Pharmaceutical Research and Development, Lundbeck, and Bristol-Myers Squibb Pharmaceuticals Limited; grants awarded by Johnson & Johnson Pharmaceutical Research & Development, Bristol-Myers Squibb Pharmaceuticals Limited, and E Merck Pharmaceuticals. All other authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Rucker, J., Breen, G., Pinto, D. et al. Genome-wide association analysis of copy number variation in recurrent depressive disorder. Mol Psychiatry 18, 183–189 (2013). https://doi.org/10.1038/mp.2011.144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2011.144
Keywords
This article is cited by
-
A sex-specific genome-wide association study of depression phenotypes in UK Biobank
Molecular Psychiatry (2023)
-
NDE1 and NDEL1 from genes to (mal)functions: parallel but distinct roles impacting on neurodevelopmental disorders and psychiatric illness
Cellular and Molecular Life Sciences (2017)
-
Translating genome-wide association findings into new therapeutics for psychiatry
Nature Neuroscience (2016)
-
Genomic structural variation in affective, anxiety, and stress-related disorders
Journal of Neural Transmission (2015)
-
Chipping away at major depressive disorder
Genome Biology (2014)