Abstract
The pattern-based classification system for HPV-related endocervical adenocarcinoma, which classifies tumors based on the destructiveness of stromal invasion, is predictive of the risk of nodal metastases and adverse outcome. Previous studies have demonstrated clinically important molecular alterations in endocervical adenocarcinoma, including KRAS and PIK3CA mutations; however, correlation between the molecular landscape and pathological variables including pattern of invasion has not been thoroughly explored. In this study, 20 endocervical adenocarcinomas were classified using the pattern-based classification system and were subjected to targeted sequencing using the Ion AmpliSeq Cancer Hotspot Panel v2 (ThermoFisher Scientific, Waltham, MA, USA) that surveys hotspot regions of 50 oncogenes and tumor suppressor genes. Single-nucleotide polymorphisms were correlated with clinical and pathologic variables including pattern of invasion. Five (25%), six (30%), and nine (45%) cases were classified as patterns A, B, and C respectively. Lymph node metastases, advanced stage at presentation and mortality from disease were exclusively seen in destructively invasive tumors (patterns B or C). Prevalent mutations in the cohort involved PIK3CA (30%), KRAS (30%), MET (15%), and RB1 (10%). Most (94%) relevant genomic alterations were present in destructively invasive tumors with PIK3CA, KRAS, and RB1 mutations seen exclusively in pattern B or C subgroups. KRAS mutations correlated with advanced stage at presentation (FIGO stage II or higher). Our findings indicate that the pattern of stromal invasion correlates with genomic abnormalities detected by next-generation sequencing, suggesting that tumors without destructive growth (pattern A) are biologically distinct from those with destructive invasion (patterns B and C), and that pattern B endocervical adenocarcinoma is more closely related to its pattern C counterpart. The pattern-based classification may be used as a triage tool when considering molecular testing for prognostic or therapeutic purposes.
Similar content being viewed by others
Main
Despite advances in prevention, diagnosis, and treatment, cancer of the uterine cervix is still the most common cancer and the leading cause of cancer-related mortality in women in 55 countries, primarily in less-developed regions of the world.1 Compared to squamous cell carcinoma, the incidence of endocervical adenocarcinoma has shown a relative increase, in part due to the difficulty of detecting glandular lesions on cytological screening.2
Infection of the cervical epithelium by oncogenic human papilloma virus (HPV) is known to be the etiological driving force behind the majority of endocervical adenocarcinomas of the usual, endometrioid and intestinal types.3 As recently demonstrated by The Cancer Genome Atlas (TCGA) Research Network, the effects of HPV in cervical carcinogenesis are diverse, and our knowledge of the activating molecular pathways in cervical adenocarcinoma is evolving.4 Prevalent mutations in cervical adenocarcinomas include PIK3CA (11–25%), KRAS (18%), and PTEN (4%) genes,5, 6, 7, 8 all members of the PI3K/Akt/mTOR signalling cascade that is important in cell cycle regulation. Importantly, tumors with these mutations may be amenable to targeted therapies.9, 10 Furthermore, presence of recurring mutations appears to be prognostic: KRAS mutations have been linked to higher rates of tumor relapse,11 whereas PIK3CA mutations have been associated with less likelihood of distant metastases.12
Early-stage (International Federation of Gynecology and Obstetrics (FIGO) stage I) endocervical adenocarcinoma is primarily treated with surgery.13 A classification of HPV-related endocervical adenocarcinoma based on the pattern of stromal invasion has been recently proposed as predictive of the risk of nodal metastasis, and is regarded as a useful tool to tailor the therapeutic approach and avoid excessive surgery in low-risk cases.14, 15, 16 Tumors with a non-destructive growth pattern (pattern A) have no risk of nodal metastases, compared to those with focal (pattern B) or diffuse (pattern C) destructive invasion (4% and 23%, respectively).16, 17 Likewise, tumor recurrence and death of disease is highly correlated with the invasive pattern.16, 18 Tumor horizontal spread, depth of invasion, and stage also correlate with pattern: in one study, 90% of pattern A tumors were stage IA, whereas 76% of pattern B and 80% of pattern C tumors were stage IB.19
The differences in morphology and behavior seen among the different patterns of invasion may reflect diverse underlying genetic mechanisms that correlate with clinical outcome. Previous studies characterizing the genomic landscape of cervical adenocarcinoma have not provided a comprehensive correlation with tumor histopathologic features, in particular the invasive pattern. Molecular profiling may explain the differences in clinical behaviour seen among different invasive patterns of cervical adenocarcinoma. Thus, we aimed to determine the prevalence of oncogene and tumor suppressor gene alterations in HPV-related endocervical adenocarcinoma and their association with prognostic pathologic variables with emphasis on the pattern of stromal invasion.
Materials and methods
This study was approved by the Research Ethics Board at Sunnybrook Health Sciences Centre.
Case Selection and Review
Cases with a diagnosis of invasive endocervical adenocarcinoma treated surgically at Sunnybrook Health Sciences Centre from 2009 to 2012 were identified through our lab information system and retrieved from our departmental archives. Non-HPV-related adenocarcinomas (gastric NOS, minimal deviation, clear cell, and serous) and cases with incompletely available histologic material or tissue blocks were excluded. Twenty cases of endocervical (usual type) adenocarcinoma with full histologic material available were selected. The diagnosis was confirmed by one gynecologic pathologist (CP-H). Histologic sections of the cervix from the 20 selected cases were independently reviewed by two gynecologic pathologists (CP-H and BD) and one pathology resident (AH), and the invasive pattern was assigned by each author. Cases with any disagreement were reviewed simultaneously by the three authors and a consensus pattern was assigned following strict criteria.17, 20
The following demographic and clinicopathologic data, obtained from the electronic patient record and pathology reports, were recorded: age at diagnosis, date and type of gynecological surgical procedure (cone biopsy, trachelectomy, hysterectomy, or exenteration), depth of invasion, horizontal extent, tumor grade, margin status, lympho-vascular invasion, number of lymph nodes sampled, number of lymph nodes involved by metastatic cervical adenocarcinoma, date of last known follow-up, and clinical status at last follow-up (alive with no evident disease, alive with metastases, dead of disease, or dead from other causes).
Genomic DNA Extraction
Samples were taken from formalin-fixed, paraffin-embedded tissue blocks. Three representative tumor areas were circled in the corresponding slide by one pathologist (CP-H) and three cores, 1 mm in width and ~3 mm in depth, were manually punched out from the respective blocks. All areas had a tumor cellularity of at least 50%. In pattern B and C adenocarcinomas, all selected tissue areas included destructively invasive tumor. The genomic DNA (gDNA) from the tissue cores was extracted by the MagMAX formalin-fixed, paraffin-embedded DNA Isolation kit (ThermoFisher). All procedures were carried out in accordance with the manufacturer’s instructions. The amount of amplifiable gDNA was determined by the TaqMan RNase P assay (ThermoFisher).
Targeted Exome Sequencing
The targeted exome sequencing was performed on the Ion S5XL next-generation sequencing (NGS) system with the Ion AmpliSeq Cancer Hotspot Panel ver.2 (CHPv2) (ThermoFisher). The panel surveys hotspot regions of 50 oncogenes and tumor suppressor genes that are commonly mutated in human cancers and covers ~2800 COSMIC mutations. The complete list of interrogated genes is listed in Table 1. The amplicon library was constructed from 10 ng of amplifiable gDNA by Ion Ampliseq Library Kit 2.0 with 207 primer pairs of the CHPv2. The targeted areas were amplified by polymerase chain reaction (PCR) for 20 cycles. The resulting amplicons were treated with FuPa reagent to partially digest primers. Amplicons were ligated to Ion P1 and Ion Xpress barcode adapters and purified using Agencourt AMPure XP reagent (Beckman Coulter, Brea, CA, USA). Barcoded libraries were quantified using the Ion Library TaqMan Quantitation Kit (ThermoFisher) and diluted to a final concentration of 40 pM. The sequencing template preparation was done using Ion Chef with Ion 520 Chef Kits. Sequencing was performed for 500 flows on an Ion S5XL Sequencer with Ion 520 chip. For detection of somatic mutations, the sequencing was performed to at least 1500-fold mean depth per sample.
Sequencing Data Analysis
The Ion Torrent platform-specific pipeline software, Torrent Suite version 5.0.5 (ThermoFisher) was used to separate barcoded reads and to filter and remove poor signal reads. BAM format files were generated from the sequencing results and then exported to the Ion Reporter Server (ThermoFisher). The bioinformatics analysis of the sequencing was performed with Ion Torrent platform-specific bioinformatics software, Ion Reporter v5.2. The Ion Reporter CHPv2 workflow was used for detection of single-nucleotide polymorphisms (SNPs), insertions or deletions (INDELs) and copy number variations (CNVs) in the tumor and for calculation of a P-value representing the statistical confidence of that call. For CNV analysis, pre-computed informatics baseline control was generated from sequencing data of CHPv2 from 20 endocervical cancers, 21 colorectal cancers and 20 melanomas. The workflow also annotates the variants with information from dbSNP, 1000 genomes, OMIM, COSMIC, Polyphen, and ClinVar.
Sanger Sequencing Validation
Mutations detected by the CHPv2 were validated by Sanger Sequencing. The nucleotide sequences of the primers in the CHPv2 were obtained from ampliseq.com and synthesized with the M13 forward primer sequence 5′-TGTAAAACGACGGCCAGT-3′ at the 5′ end of the forward primer sequence and the M13 reverse primer sequence 5′- CAGGAAACAGCTATGACC-3′ at the 5′ end of the reverse primer (Integrated DNA Technologies, Coralville, IA, USA). One nanogram of formalin-fixed, paraffin-embedded DNA was used for amplification of target region and cycle-sequencing reaction was performed with the BigDye Direct Cycle Sequencing Kit (ThermoFisher). The resulting sequencing reaction was purified by the BigDye XTerminator Purification Kit (ThermoFisher) and automated sequencing was performed by capillary electrophoresis on an ABI3500 (Applied Biosystems) with POP7 polymer. The sequencing data was analyzed by FinchTV ver1.4.0 (Geospiza).
Immunohistochemistry
Immunohistochemistry for p53 (clone DO7, prediluted, Ventana, Tucson, AZ, USA) and Her2Neu (HercepTest, DAKO, Carpinteria, CA, USA) was performed on paraffin-embedded tumor sections. p53 was interpreted as normal (patchy, heterogeneous nuclear staining) or abnormal (strong nuclear expression in ≥80% of cells, or complete absence of staining). Her2Neu was interpreted as negative (absent staining), 1+ (weak, incomplete staining), 2+ (complete weak membranous staining in at least 10% of cells or complete intense staining in <30% of the cells), and 3+ (complete intense membranous staining in >30% of the cells).
Statistical Analysis
Inferential analysis was performed to compare clinicopathologic variables and genomic data between pattern subgroups using GraphPad QuickCalcs (GraphPad Software, La Jolla, CA, USA); a P-value of <0.05 was considered statistically significant.
Results
Review of Clinical and Pathologic Data
Clinical and pathologic data are summarized in Table 2. The mean age at diagnosis was 43 years (median 39 years, range 29–80 years). The majority of the included patients were treated with hysterectomy (n=13, 65%); the remaining patients underwent trachelectomy (n=3, 15%), cone biopsy (n=2, 10%), or pelvic exenteration (n=2, 10%). The mean duration of follow-up at our institution was 38 months (median 46 months, range 1–77 months). One patient (case 14) died of her disease; all other patients were alive with no evidence of recurrence at their last follow-up.
Pattern of Invasion Classification and Correlation with Pathologic Features
Five (25%), six (30%), and nine (45%) tumors were classified as patterns A, B, and C, respectively (Figure 1). Fifteen cases (75%) had full agreement on pattern assignation by three independent reviewers. In the remaining five, agreement was reached between two reviewers, with a discordant pattern assignment by the third reviewer. In two of these, initial disagreement was between patterns B and C. In the remaining three cases (15%), the discordance was between pattern A and pattern B or C. In all five tumors, initial disagreement on pattern assignation was fully resolved upon consensus review.
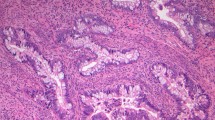
Classification of endocervical adenocarcinoma by pattern of invasion. (a, b) Pattern A adenocarcinoma: well-demarcated glands with rounded contours and lack of destructive growth or lymphovascular space invasion. (c, d) Pattern B adenocarcinoma: irregular glands and cell clusters arising from pattern A glands with lack of solid growth or marked complexity. (e, f) Pattern C adenocarcinoma: diffusely infiltrative tumor with angulated glands, extensive desmoplastic response, and confluent growth.
None of the patients with pattern A or B tumors that underwent lymphadenectomy had nodal tumor spread; conversely, 33% (3/9) of patients with pattern C tumors had lymph node metastases (P=0.07). All (100%) pattern A and four (67%) pattern B tumors were FIGO stage IB. Conversely, only five (56%) pattern C tumors were stage IB, whereas two (22%) were stage IIIB based on nodal involvement, and two (22%) were stage IV. The two pattern C tumors with stage IIIB based on nodal involvement had otherwise no other features warranting advanced stage (by size alone would be categorized as IB). All pattern A cases were grade 1, while grade 3 cases were exclusively classified as pattern C; grade 2 cases (n=8) were classified as pattern B (n=3, 38%) or pattern C (n=5, 63%) (P<0.001). The pattern of invasion did not correlate significantly with depth of invasion, horizontal extent, or margin status. The presence of lymphovascular invasion is an exclusionary criterion for pattern A designation, and therefore was not statistically analyzed.
Genomic Alterations in Endocervical Adenocarcinoma
Seventeen prevalent mutations were detected, with an average of 1.45 alterations/case (Table 3). Missense mutations were identified in PIK3CA (six cases, 30%), KRAS (six cases, 30%), MET (three cases, 15%), and RB1 (two cases, 10%). Sanger sequencing confirmed these mutations in all cases except for two (peak present, but lower than limit of detection). PIK3CA and KRAS mutations were concurrently present in four cases. Nonsense or frameshift mutations were not identified.
Copy number gains ≥4 were seen with ERBB2 (six cases, 30%), TP53 (four cases, 20%), and HNF1A (two cases, 10%). Two-copy (homozygous) losses were not identified. Interestingly, none of the tumors with ERBB2 copy gains showed Her2Neu overexpression by immunohistochemistry (all had a score of 0). Likewise, none of the tumours with TP53 copy gains demonstrated an abnormal p53 expression pattern.
Correlation Between Genomic Abnormalities and Pathologic Features
The distribution of SNPs among cases classified by pattern is depicted in Figure 2. Overall, mutations were more frequent in destructively invasive adenocarcinoma with an average of 1.2 mutations per case in pattern A tumors compared with 2.3 mutations per case in pattern B and C tumors (P=0.04). Out of the relevant mutations identified, only one (6%) was identified in the pattern A group compared to nine (53%) corresponding to pattern B and seven (41%) to pattern C; the difference in mutation prevalence between non-destructive (pattern A) and destructively invasive (patterns B and C) tumors was significant (P=0.0001). Five out of six (83%) pattern B and five out of nine (56%) pattern C cases harbored at least one gene alteration; in contrast, the same was observed in only one pattern A tumor (20%).
Mutations in PIK3CA and KRAS were seen exclusively in destructively invasive tumors (P=0.05): 4/6 (67%) pattern B tumors and 4/9 (44%) pattern C tumors. All mutations involving these genes are listed in the Sanger Institute Catalogue Of Somatic Mutations In Cancer (COSMIC) web site, http://cancer.sanger.ac.uk/cosmic.21 In addition, novel RB1 mutations were identified in two cases (one pattern B and one pattern C), and MET mutations in three additional cases (one pattern A and two pattern B). Presence of KRAS mutation was associated with advanced stage: 4/6 patients with FIGO stage II or greater harbored KRAS mutations, vs 2/14 patients with FIGO stage I (P=0.04). None of the remaining SNPs identified were statistically associated with depth of invasion, lymphovascular invasion, lymph node metastases, or FIGO stage (IB vs II–IV).
ERBB2 copy number gains (greater than four copies) were seen in 1/5 (20%), 2/6 (33%), and 3/9 (30%) pattern A, B, and C cases, respectively; they occurred synchronously with KRAS and PIK3CA mutations in 4/5 (80%) destructively invasive tumors. Concurrent TP53 gain was seen in 4/6 (67%) cases with ERBB2 gains, all of which were destructively invasive (one pattern B and three pattern C tumors). HNF1A copy number gains were observed in two tumors (10%, both pattern C).
Discussion
The histopathologic approach to patients with HPV-related endocervical glandular neoplasia has recently evolved toward a combination of standardly reported metrics (such as tumor size) and morphologic assessment of the invasive pattern as per the pattern-based classification system. Although our cohort is relatively small, our findings support the mounting body of evidence suggesting that the pattern of stromal invasion is a useful indicator of a patient’s risk of nodal metastasis, stage, and now the presence of oncogenic mutations.
The goal of this study was to identify prevalent oncogenic mutations in endocervical adenocarcinoma and to correlate these alterations with important pathologic variables. For the first time, we present an important relationship between pattern of invasion and specific oncogenic mutations, some of which are or may be therapeutically targetable in the future.
We found mutations in PIK3CA and KRAS to be prevalent in our cohort (30% each), in keeping with previously reported rates including The Cancer Genome Atlas data.4 KRAS and PIK3CA are critical components of the PI3K/MAPK pathway, which illustrates the potential for targeted therapeutic agents in cervical adenocarcinoma.10, 22 Remarkably, mutations in these two genes were identified exclusively in destructively invasive tumors (patterns B and C). In this context, a pattern-based approach may serve as a tool to identify cases for molecular testing and, potentially, personalized therapy. Moreover, KRAS and PIK3CA mutations in endocervical adenocarcinoma have potential prognostic importance, and may explain the aggressive biologic potential of pattern B and C tumors. An association between KRAS and destructive invasion has been observed in endometrial carcinoma, in which KRAS mutations are frequent in tumors with a microcystic, elongated and fragmented (MELF) pattern of invasion.23 Moreover, KRAS mutations in our cohort were significantly associated with advanced stage at presentation (FIGO stage II or greater). In a separate study, KRAS mutations were associated with higher rates of tumor relapse.11 PIK3CA mutations appear to occur late in cervical carcinogenesis according to previous data on cervical squamous neoplasia.24 This observation may explain the strong correlation between destructively invasive adenocarcinoma (particularly pattern C) and PIK3CA mutation status.
Mutations in KRAS and PIK3CA found in our cohort have been previously recorded in human malignancies and can be found in the COSMIC database. We also identified mutations in RB1 and MET, not previously reported in COSMIC. RB1, which encodes to the tumor suppressor retinoblastoma protein (pRb), has been previously implicated in the carcinogenesis of many different malignancies. Alterations in pRb expression are known in HPV-related tumours including cervical carcinomas, as the viral E7 oncoprotein downregulates the functionality of this important tumor suppressor.25 Direct inactivation of the RB1 gene by mutation has been described in different malignancies such as childhood retinoblastoma26 and small cell lung carcinoma,27 as well as in HPV-negative cervical carcinoma cell lines. Presumably, missense mutations in the RB1 gene cause reduced functionality in the pRb protein, analogous to the effect the E7 oncoprotein has, albeit through a different mechanism. Mutations in the MET gene, which encodes the tyrosine kinase hepatocyte growth factor receptor (HGFR), have been frequently reported in various malignancies;28 in our study, we detected MET mutations in 15% of tumours, which represents a novel finding. Importantly, the HGFR tyrosine kinase is under investigation as a oncologic therapeutic target.29, 30, 31 Finally, single-nucleotide substitutions involving TP53 have previously been reported in up to 36% of endocervical adenocarcinomas,8 a finding not observed in our study.
Copy number variations were identified across all pattern categories, but were considerably more common in destructively invasive tumors compared to non-destructive tumors. Alterations in the ERBB2 gene, which encodes the human epidermal growth factor receptor 2 (Her2Neu), have been reported in different malignancies32, 33 including cervical carcinomas.5, 34 Furthermore, ERBB2 amplification has been shown to correlate with poor prognosis in endocervical adenocarcinoma,35 especially in those with lymph node metastases.36 ERBB2 copy number gains were identified in six cases (30%), five of which were pattern B or C. Importantly, there were concurrent gains in TP53 in four of these tumors, which highly suggest chromosome 17 polysomy.37 The only pattern A case harboring ERBB2 gains displayed confluent growth suggestive of pattern C; however, it was mostly seen in the surface and was <5 mm in diameter (thus, warranting classification as pattern A). It is possible that ERBB2 gains represent an early genomic event not yet translated into a frankly destructively invasive pattern. We performed immunohistochemistry for p53 and Her2Neu to determine whether the presence of ERBB2 and TP53 gains correlated with increased protein expression. Interestingly, the copy gains in ERBB2 and TP53 did not translate into Her2Neu or p53 protein overexpression, which may be explained by the relative low copy number gain observed (>4 but <10). The lack of perfect correlation between gene amplification and protein overexpression has previously been demonstrated for ERBB2 in non-cervical malignancies,38, 39 and thus deserves further study.
The homeobox family protein hepatocyte nuclear activator 1-alpha, encoded by HNF1A, is a transcriptional activator necessary for the tissue-specific expression of many genes. Defects in HNF1A have been linked with the development of mature onset diabetes of the young type 3 (MODY 3)40 as well as hepatic adenomas.41 Recently, its role in pancreatic carcinogenesis is under investigation.41 Neither mutations nor copy number gains in HNF1A have been previously linked to adenocarcinoma of the cervix. Our observation of HNF1A gain in 10% of our cases may suggest an additional novel alteration in destructively invasive adenocarcinomas, as HNF1A copy number gains were identified exclusively in pattern C tumours in our cohort.
Overall, out of the relevant genomic alterations identified, the vast majority corresponded to destructively invasive tumors (patterns B and C). This suggests that pattern A tumors are biologically distinct from pattern B and C tumors. From this perspective, pattern A adenocarcinoma may represent: (a) an early step in the progression of endocervical adenocarcinoma in which oncogenic mutations occur subsequently conferring tumor aggressiveness, or (b) a distinct, indolent subset of HPV-related endocervical glandular neoplasia with inherent lack of oncogenic alterations.
We did not observe significant differences between patterns B and C in respect to distribution of oncogenic abnormalities. However, KRAS mutations appeared more frequently in pattern B tumors, whereas PIK3CA mutations and gene copy number variations were more common in pattern C tumors. These observations require further study. The genomic similarities between these two patterns suggest that they are biologically related, and likely represent a more aggressive form of endocervical adenocarcinoma. Indeed, pattern B and C tumors are relatively similar in terms of tumor size and stage, and their distinction suffers from significant inter-observer variability.18, 20
Appraisal of the biologic and clinical significance of the pattern-base classification requires consideration of the inter-observer reproducibility of pattern assignation. As demonstrated previously, the inter-observer agreement of the classification is adequate, and improves if simplified to a two-tier system (non-destructive vs destructive invasion).18, 20, 42 Pattern in all cases in our cohort had complete agreement among three authors, two of which have considerable experience in the study of the pattern-based classification. As such, we consider that the potential impact of inter-observer variation in our analysis was minimized.
Our study was limited by its relatively small sample size and by the short follow-up period in some patients, because their presentation was recent or they were lost in follow-up. In addition, it is important to note that our cohort included a subset of destructively invasive tumors (three pattern C cases) with no mutations of CNVs identified; one of these progressed to distant metastatic disease ultimately leading to the patient’s demise. Other genomic abnormalities not surveyed by our platform of choice may account for these cases. Therefore, further studies with more comprehensive genomic sequencing platforms are necessary.
In summary, our study revealed a significantly higher prevalence of oncogene and tumor suppressor gene abnormalities in pattern B and C invasive endocervical adenocarcinomas. On the basis of the genomic profile, non-destructive forms of adenocarcinoma appear to be biologically different from destructive ones: pattern A lesions may constitute early phases of tumor progression or true distinct indolent forms of adenocarcinoma, whereas patterns B and C represent a more aggressive subset of cervical glandular neoplasia. In concert with other prognostic variables, the pattern-based classification may serve as a triage tool for molecular testing in order to assess prognosis and to identify patients amenable to targeted treatments.
References
World Health Organization. Comprehensive Cervical Cancer Control: A Guide to Essential Practice, 2nd edn. [Internet]. World Health Organization Press: Geneva, 2014, p 27-28.
Smith HO, Tiffany MF, Qualls CR et al. The rising incidence of adenocarcinoma relative to squamous cell carcinoma of the uterine cervix in the United States—a 24-year population-based study. Gynecol Oncol 2000;78:97–105.
Pirog EC, Kleter B, Olgac S et al. Prevalence of human papillomavirus DNA in different histological subtypes of cervical adenocarcinoma. Am J Pathol 2000;157:1055–1062.
Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017;543:378–384.
Ojesina AI, Lichtenstein L, Freeman SS et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014;506:371–375.
Wright AA, Howitt BE, Myers AP et al. Oncogenic mutations in cervical cancer: genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer 2013;119:3776–3783.
Lou H, Villagran G, Boland JF et al. Genome analysis of Latin American cervical cancer: frequent activation of the PIK3CA pathway. Clin Cancer Res 2015;21:5360–5370.
Tornesello ML, Annunziata C, Buonaguro L et al. TP53 and PIK3CA gene mutations in adenocarcinoma, squamous cell carcinoma and high-grade intraepithelial neoplasia of the cervix. J Transl Med 2014;12:255.
Fruman DA, Rommel C . PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 2014;13:140–156.
Chappell WH, Steelman LS, Long JM et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget 2011;2:135–164.
Xiang L, Li J, Jiang W, Shen X, Yang W, Wu X et al. Comprehensive analysis of targetable oncogenic mutations in chinese cervical cancers. Oncotarget 2014;6:4968–4975.
Xiang L, Jiang W, Li J, Shen X, Yang W, Yang G et al. PIK3CA mutation analysis in Chinese patients with surgically resected cervical cancer. Sci Rep 2015;5:14035.
National Cancer Institute. Cervical Cancer Treatment. Available at https://www.cancer.gov/types/cervical/hp/cervical-treatment-pdq#section/_85.
Pecorelli S, Zigliani L, Odicino F . Revised FIGO staging for carcinoma of the cervix. Int J Gynaecol Obstet 2009;105:107–108.
Roma AA . Patterns of invasion of cervical adenocarcinoma as predicators of outcome. Adv Anat Pathol 2015;22:345–354.
Roma AA, Diaz De Vivar A, Park KJ et al. Invasive endocervical adenocarcinoma: a new pattern-based classification system with important clinical significance. Am J Surg Pathol 2015;39:667–672.
Diaz De Vivar A, Roma AA, Park KJ et al. Invasive endocervical adenocarcinoma: proposal for a new pattern-based classification system with significant clinical implications: a multi-institutional study. Int J Gynecol Pathol 2013;32:592–601.
Parra-Herran C, Taljaard M, Djordjevic B et al. Pattern-based classification of invasive endocervical adenocarcinoma, depth of invasion measurement and distinction from adenocarcinoma in situ: interobserver variation among gynecologic pathologists. Mod Pathol 2016;29:879–892.
Djordjevic B, Parra-Herran C . Application of a pattern-based classification system for invasive endocervical adenocarcinoma in cervical biopsy, cone and loop electrosurgical excision (LEEP) material: pattern on cone and LEEP is predictive of pattern in the overall tumor. Int J Gynecol Pathol 2016;35:456–466.
Rutgers JKL, Roma AA, Park KJ et al. Pattern classification of endocervical adenocarcinoma: reproducibility and review of criteria. Mod Pathol 2016;29:1083–1094.
Bamford S, Dawson E, Forbes S et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 2004;91:355–358.
Hou M-M, Liu X, Wheler J et al. Targeted PI3K/AKT/mTOR therapy for metastatic carcinomas of the cervix: a phase I clinical experience. Oncotarget 2014;5:11168–11179.
Stewart CJR, Amanuel B, Grieu F et al. KRAS mutation and microsatellite instability in endometrial adenocarcinomas showing MELF-type myometrial invasion. J Clin Pathol 2010;63:604–608.
Verlaat W, Snijders PJ, van Moorsel MI et al. Somatic mutation in PIK3CA is a late event in cervical carcinogenesis. J Pathol Clin Res 2015;1:207–211.
Yim E-K, Park J-S . The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat 2005;37:319–324.
Ali MJ, Parsam VL, Honavar SG et al. RB1 gene mutations in retinoblastoma and its clinical correlation. Saudi J Ophthalmol 2010;24:119–123.
George J, Lim JS, Jang SJ et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53.
Zenali M, deKay J, Liu Z et al. Retrospective review of MET gene mutations. Oncoscience 2015;2:533–541.
Scagliotti GV, Novello S, von Pawel J . The emerging role of MET/HGF inhibitors in oncology. Cancer Treat Rev 2013;39:793–801.
Kim ES, Salgia R. MET . MET pathway as a therapeutic target. J Thorac Oncol 2009;4:444–447.
Naran S, Zhang X, Hughes SJ . Inhibition of HGF/MET as therapy for malignancy. Expert Opin Ther Targets 2009;13:569–581.
Scholl S, Beuzeboc P, Pouillart P . Targeting HER2 in other tumor types. Ann Oncol 2001;12:S81–S87.
English DP, Roque DM, Santin AD . HER2 expression beyond breast cancer: therapeutic implications for gynecologic malignancies. Mol Diagn Ther 2013;17:85–99.
Yan M, Parker BA, Schwab R et al. HER2 aberrations in cancer: implications for therapy. Cancer Treat Rev 2014;40:770–780.
Kihana T, Tsuda H, Teshima S et al. Prognostic significance of the overexpression of c-erbB-2 protein in adenocarcinoma of the uterine cervix. Cancer 1994;73:148–153.
Hale RJ, Buckley CH, Fox H et al. Prognostic value of c-erbB-2 expression in uterine cervical carcinoma. J Clin Pathol 1992;45:594–596.
Tse CH, Hwang HC, Goldstein LC et al. Determining true HER2 gene status in breast cancers with polysomy by using alternative chromosome 17 reference genes: implications for anti-HER2 targeted therapy. J Clin Oncol 2011;29:4168–4174.
Slamon DJ, Godolphin W, Jones LA et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989;244:707–712.
Kakar S, Puangsuvan N, Stevens JM et al. HER-2/neu assessment in breast cancer by immunohistochemistry and fluorescence in situ hybridization: comparison of results and correlation with survival. Mol Diagn J 2000;5:199–207.
Bellanné-Chantelot C, Carette C, Riveline J-P et al. The type and the position of HNF1A mutation modulate age at diagnosis of diabetes in patients with maturity-onset diabetes of the young (MODY)-3. Diabetes 2008;57:503–508.
Jeannot E, Mellottee L, Bioulac-Sage P et al. Spectrum of HNF1A somatic mutations in hepatocellular adenoma differs from that in patients with MODY3 and suggests genotoxic damage. Diabetes 2010;59:1836–1844.
Paquette C, Jeffus SK, Quick CM et al. Interobserver variability in the application of a proposed histologic subclassification of endocervical adenocarcinoma. Am J Surg Pathol 2015;39:93–100.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hodgson, A., Amemiya, Y., Seth, A. et al. Genomic abnormalities in invasive endocervical adenocarcinoma correlate with pattern of invasion: biologic and clinical implications. Mod Pathol 30, 1633–1641 (2017). https://doi.org/10.1038/modpathol.2017.80
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.80
This article is cited by
-
Silva cumulative score and its relationship with prognosis in Endocervical adenocarcinoma
BMC Cancer (2022)
-
Genomic alterations caused by HPV integration in a cohort of Chinese endocervical adenocarcinomas
Cancer Gene Therapy (2021)
-
Clinical implication of oncogenic somatic mutations in early-stage cervical cancer with radical hysterectomy
Scientific Reports (2020)
-
The pattern is the issue: recent advances in adenocarcinoma of the uterine cervix
Virchows Archiv (2018)