Abstract
Approximately 1–2% of pancreatic neoplasms are acinar cell carcinomas. Recently, BRAF gene rearrangements were identified in over 20% of acinar-type neoplasms, which included both pure acinar cell carcinomas and mixed carcinomas with acinar differentiation, using next-generation sequencing-based platforms, providing a potential therapeutic target for patients with these neoplasms. Thus, it is clinically important to develop a rapid, cost- and material-efficient assay to screen for BRAF gene fusions in pancreatic acinar-type neoplasms. We developed a dual color, break-apart FISH assay to detect BRAF gene rearrangements in these neoplasms and evaluated its performance in comparison to next-generation sequencing-based studies. A blinded BRAF rearrangement FISH investigation was performed on 31 acinar-type neoplasms that had been studied previously by next-generation sequencing-based analysis as well as on 18 additional acinar-type neoplasms that were accrued over the past 2 years. In total, BRAF fusions were identified in 12/49 (24%) acinar-type neoplasms by FISH. BRAF fusion partners were uncovered by using targeted next-generation sequencing studies in 11 FISH-positive cases that had sufficient material for next-generation sequencing studies. SND1 was the most frequent fusion partner involved in BRAF-fusion acinar-type neoplasms (50%), followed by HERPUD1 (18%). No BRAF fusions were identified by next-generation sequencing in any of the FISH-negative cases investigated. FISH analysis showed that BRAF rearrangements were diffusely present across tumor-rich areas in BRAF-fusion acinar-type neoplasms, which is consistent with an oncogenic driver alteration pattern. Thus, we demonstrated that, in comparison to targeted next-generation sequencing-based technologies, the FISH assay is highly sensitive and specific as well as time- and cost-efficient in the detection of BRAF fusions in acinar-type neoplasms. The FISH assay can be easily implemented in diagnostic settings to identify acinar-type neoplasms patients potentially suitable for targeted therapy to inhibit MAPK pathway activity.
Similar content being viewed by others
Main
Pancreatic acinar cell carcinomas account for 1–2% of pancreatic cancers.1 In addition to characteristic morphological features, the diagnostic hallmark of acinar cell carcinomas is the immunohistochemical expression of acinar-specific exocrine enzymes, such as trypsin and chymotrypsin.2 The common genetic aberrations of ductal adenocarcinomas, such as mutations in KRAS, DPC4, and TP53 genes, are generally absent or uncommon in pancreatic acinar-type neoplasms. Alterations in the APC/β-catenin pathway, either through activating mutations of CTNNB1 gene or truncating mutations of APC gene, have been reported in a subset of pancreatic acinar-type neoplasms (20–50%).2 As chemo and radiation therapies have limited efficacy against these tumors, exploring genetic aberrations in pancreatic acinar-type neoplasms to identify actionable molecular targets may trigger the development of novel therapies.
In a recent study, our group performed comprehensive genomic profiling of 44 pancreatic acinar-type neoplasms (including mixed carcinomas with acinar differentiation) using a next-generation sequencing-based technical platform and identified BRAF gene rearrangements in over 20% of the neoplasms.3 The pilot next-generation sequencing study revealed eight different BRAF fusions, five of which were SND1–BRAF with minor differences in the fusion points. Despite a diversity of fusion partners and fusion points, all BRAF rearrangements involved relatively conserved BRAF break points that preserve the kinase domain. The pilot study also demonstrated that the most prevalent fusion in pancreatic acinar-type neoplasms, SND1–BRAF, was oncogenic through the activation of the MAPK pathway, which could be abrogated in vitro with MEK inhibition. The identification of BRAF fusions in a significant subset of pancreatic acinar-type neoplasms provides a potential therapeutic target for patients with this type of tumor. Thus, it is clinically important to develop a rapid, cost- and material-efficient assay to screen for BRAF gene fusions in acinar-type neoplasms.
Given that multiple fusion partners and variable fusion points challenge RT-PCR primer design, we developed a dual-color break-apart gene rearrangement FISH assay. In this study, we evaluated the FISH assay for the detection of BRAF fusions in acinar-type neoplasms with a comparison to next-generation sequencing-based studies.
Materials and methods
Study Population
This study was conducted with the approval of the Memorial Sloan Kettering Cancer Center Institutional Review Board. The study included both pure acinar cell carcinomas and mixed carcinomas with acinar differentiation, such as mixed acinar neuroendocrine carcinoma, mixed acinar ductal carcinoma, and mixed acinar neuroendocrine ductal carcinoma, all of which exhibit predominantly acinar differentiation and are known to be pathologically related to pure acinar cell carcinoma;4 for simplicity, all cases are collectively referred to as acinar-type neoplasms henceforth. We included 31 acinar-type neoplasms that were previously used in the pilot study (discovery cohort) for the discovery of BRAF fusions and an independent cohort (second cohort) of 18 acinar-type neoplasms cases that were accrued over the past 2 years. The entire study set of 49 acinar-type neoplasms included 45 pancreatic and 4 hepatic primaries, in 35 males and 13 females, with a median age of 59 (range of 15–80 years old). Demographics and specific histopathological diagnosis for each case are presented in Supplementary Table 1.
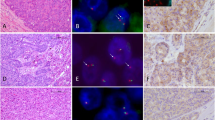
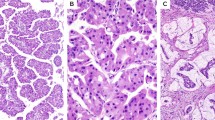
The discovery cohort included 17 pure acinar cell carcinomas, 7 mixed acinar neuroendocrine carcinomas, 4 mixed acinar ductal carcinomas, and 3 mixed acinar neuroendocrine ductal carcinomas. The discovery cohort, in which BRAF fusions were identified in six cases using a next-generation sequencing-based approach, was used to validate the FISH assay. The second cohort, used to further evaluate the prevalence of BRAF fusions in acinar-type neoplasms, included nine pure acinar cell carcinomas (three were hepatic primaries), six mixed acinar neuroendocrine carcinomas, one mixed acinar ductal carcinoma (also a hepatic primary), and two mixed acinar neuroendocrine ductal carcinomas. Representative hematoxylin and eosin images of acinar-type neoplasms in our series are presented in Figure 1.
Representative images of acinar-type carcinomas in our series. (a, b) Pure acinar cell carcinomas revealed several different architectural patterns: (a) the solid pattern; (b) the acinar; (c) one of the four hepatic primary cases; (d) acinar differentiation was confirmed with trypsin immunohistochemical staining for each case; (e) the vast majority of mixed acinar neuroendocrine carcinomas were composed of a morphologically homogenous population of cells and the divergent differentiation could not be detected without immunohistochemical labeling. However, in some cases, there were two distinct populations of neoplastic cells, one with acinar features and the other having a neuroendocrine appearance as shown here; (f) in mixed acinar ductal carcinomas, glandular structures were also usually superimposed on the solid architecture of conventional acinar cell carcinomas.
BRAF Break-Apart FISH Assay
Formalin-fixed paraffin-embedded tissue samples from all 49 cases were tested for BRAF rearrangements by a break-apart FISH assay using a commercial BRAF break-apart FISH probe (Empire Genomics), which is a mixture of two clone contigs hybridizing to the chromosome band 7q34 flanking the BRAF gene locus. The green fluorochrome-labeled probe hybridizes telomeric (5′) to BRAF; the orange fluorochrome-labeled probe hybridizes centromeric (3′) to BRAF. Four-micron (4 μm) tissue sections generated from formalin-fixed paraffin-embedded blocks of tumor specimens were pretreated by deparaffinizing in xylene and dehydrating in ethanol. Dual-color FISH assays were conducted according to the protocol for formalin-fixed paraffin-embedded sections from Vysis/Abbott Molecular with minor adjustments of pepsin treatment as needed. FISH analysis and signal capture were conducted on a fluorescence microscope (Zeiss) coupled with the ISIS FISH Imaging System (Metasystems). We analyzed 100 interphase nuclei from tumor-rich areas in each specimen.
Similar to the FDA-approved ALK FISH assay, BRAF rearrangements may appear as either ‘broken apart’ signals (red and green signals separated by a gap) or a ‘single orange (3′)’ signal (missing green signal), which indicates the loss of 5′ BRAF but preservation of the 3′ BRAF coding the kinase domain. For the ‘broken apart’ signal pattern, red and green signals separated by a gap larger than 2 signal diameters were considered diagnostically positive, which is identical to the diagnostic criteria for the ALK-rearrangement FISH assay.
DNA-Based Targeted Next-Generation Sequencing
For the 18 new cases recruited from June 2014 to May 2016, 14 patients receiving treatment at Memorial Sloan Kettering Cancer Center were consented for molecular testing with Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), a capture-based next-generation sequencing assay that detects somatic single-nucleotide mutations, small indels, and copy number alterations in 410 cancer-related genes, and selected structural rearrangements of 14 recurrently rearranged genes, including BRAF, in solid tumors (MSK-IMPACT, detailed methods have been previously published).5 Briefly, 100–250 ng DNA extracted from formalin-fixed paraffin-embedded tissue with >10% tumor content and matched normal DNA from blood samples, if available, were obtained for testing with MSK-IMPACT.
Anchored Multiplex RT-PCR and Next-Generation Sequencing
Tumor-rich areas were macro-dissected from sections of formalin-fixed paraffin-embedded tissues, and subjected to RNA extraction using Mineral Oil-Qiagen RNeasy formalin-fixed paraffin-embedded kit (Qiagen, USA). Random priming was used to synthesize cDNAs from the RNA samples extracted. An anchored multiplex PCR assay based on the Rapid Amplification of cDNA Ends (RACE) strategy followed by next-generation sequencing (Archer FusionPlex, ArcherDX, USA) was employed to identify-specific partners involved in selected gene fusions.6 We used a custom fusion-screening panel focused on 35 genes, including BRAF, known to be involved in recurrent gene fusions in solid tumors. The list of genes in the panel is presented in the Supplementary Table 2. Within this panel, for BRAF fusions, 5′ RACE was conducted using BRAF-specific 3′ primers targeting exons 7–12, and the multiplex RACE products were sequenced on MiSeq (Illumina), and sequencing data were analyzed by Archer software (version 3.3.0).
Immunohistochemical Staining
Immunolabeling for phospho-ERK was performed on tissue sections of each tumor using a rabbit monoclonal antibody (Cell Signaling Technology), phospho-p44/42MAPK (202Y284). The primary antibody was applied at a dilution of 1:1000 with an incubation time of 60 min. The secondary biotinylated anti-rabbit antibody was applied at a dilution of 1:200 for 60 min, and diamino benzidine was used as the chromagen. Staining was performed using DISCOVERY DAB Map Detection Kit (Ventana) and the Ventana Discovery XT automated immunohistochemical staining platform. The immunolabeled slides were scored as previously described.3 In brief, labeling of ≥25% of nuclei and cytoplasm was scored as ‘positive’; labeling of <25% of nuclei and cytoplasm was interpreted as ‘focal’, and no staining was considered ‘negative’.
Statistical Analysis
All statistical tests were performed using the IBM-SPSS version 20.0, and the threshold for statistical significance was set at P<0.05. Data were analyzed using a Chi-square and Fisher exact tests to compare the BRAF fusion status with age, gender, tumor size, lymphovascular invasion, tumor stage (pT), and lymph node stage (pN). Survival data were analyzed by using the Kaplan–Meier method and were assessed by log-rank test to compare differences in survival between the patients whose tumors revealed BRAF fusions and those whose tumors did not reveal BRAF fusions.
Results
Development of BRAF FISH and New Patient Screening
To validate the dual-color break-apart BRAF rearrangement FISH assay, a blinded FISH investigation was performed on 31 acinar-type neoplasms from the discovery cohort, among which, BRAF fusions were previously identified in six cases by next-generation sequencing-based methods. FISH identified BRAF gene rearrangements in seven acinar-type neoplasms (23%) in the discovery cohort, including all six cases positive for BRAF fusions in the prior next-generation sequencing-based studies and one additional case that had not been identified by next-generation sequencing (case #1_10, Table 1 and Supplementary Table 1). FISH analysis was repeated in this sample, and in both assays bona fide BRAF gene rearrangement was observed in a high percentage of cells (~90%). Considering the limited technical sensitivity of targeted DNA-seq in the detection of gene fusions, we performed targeted RNA-seq (Archer FusionPlex) to further investigate this case; however, RNA-seq failed due to suboptimal sample quality. The validated BRAF rearrangement FISH assay was then employed to screen for BRAF fusions in 18 newly accrued acinar-type neoplasms (second cohort), and 5 additional BRAF-rearranged cases were identified (28%). Representative FISH images are presented in Figure 2.
Interphase FISH detection of BRAF rearrangement in arcina-type carcinomas. Representative images for each FISH signal patterns. (a) FISH negative for BRAF rearrangement, showing fusion signals of 5′ and 3′ BRAF; (b) FISH positive for BRAF rearrangement—‘broken apart’ signal pattern (red and green signals separated by a gap); (c) FISH positive for BRAF rearrangement—‘single orange (3′BRAF)’ signal pattern, which indicates the loss of 5′ BRAF but preservation of the 3′ BRAF coding the kinase domain.
In total, we identified BRAF fusions in 12 of 49 acinar-type neoplasms (24%). The frequency of BRAF rearrangement identified in this expanded study is similar to that in the prior study. Table 1 summarizes the percentage of positive cells and the dominant FISH signal pattern for each of the 12 BRAF-rearranged samples. The specific histologic subtypes (pure acinar cell carcinoma, mixed acinar neuroendocrine, mixed acinar ductal, or mixed acinar neuroendocrine ductal carcinomas), BRAF fusion partners and phosphorylated ERK (pERK) immunohistochemical staining results are shown in Table 1 as well. In all except one (11/12) BRAF-rearranged case, positive signals were observed in 60–100% of the cells analyzed (Avg=84%) in tumor-rich areas. The one ‘outlier’ case (#1_25, Table 1 and Supplementary Table 1), which harbored an SND1–BRAF fusion, showed positivity in 38% of the cells, according to our stringent FISH signal scoring criteria. However, many cells that were not considered diagnostically positive by the scoring criteria had ‘broken apart’ red and green signals separated by 1.5–2 signal diameters, which fell below the threshold for positivity. SND1 and BRAF are ~13 Mb apart on genomic DNA, respectively mapping to 7q32 and 7q34, and running in opposite directions on chromosome 7. We suspect a complex chromosomal rearrangement occurred at 7q in this sample.
Comparison of FISH to Next-generation Sequencing-Based Studies in the Detection of BRAF Fusion
BRAF fusion was also explored by the targeted DNA-seq (MSK-IMPACT) assay in 14 of 18 acinar-type neoplasms in the second cohort, including all FISH-positive cases (five cases) and nine FISH-negative cases. MSK-IMPACT identified BRAF fusions in four cases, and the remaining FISH-positive case was further analyzed by the targeted RNA-seq (Archer), and an HERPUD1–BRAF fusion was identified. It is worth noting that, besides the strong evidence from MSK-IMPACT or Archer showing bona fide BRAF fusions, no other gene fusion was identified in any of the five FISH-positive samples. Targeted RNA-seq (Archer) was also used to analyze two of four FISH-negative cases that could not be analyzed by MSK-IMPACT, and no BRAF fusions were identified. The remaining two BRAF FISH-negative cases had insufficient tumor tissue for targeted DNA-seq or RNA-seq studies. As comprehensive mutation profiling of acinar-type neoplasms has been presented previously by our group and others,3, 7 a complete genetic aberration analysis was not performed on the next-generation sequencing data obtained from the second cohort cases in the present study; instead, we focused on BRAF-related genetic alterations only. No mutation in BRAF or RAS was identified in any of the samples in the second cohort.
In our expanded study, SND1 was still the most frequent fusion partner involved in BRAF-fusion acinar-type neoplasms (50%), followed by HERPUD1 (2/11) (Table 1). All BRAF fusion partners in acinar-type neoplasms, except SND1, are located on chromosomes other than 7. A summary of findings in all 49 acinar-type neoplasms by FISH and next-generation sequencing-based studies is available in Supplementary Table 1. In brief, using next-generation sequencing-based studies (targeted DNA-seq or RNA-seq), in-frame BRAF fusions were identified in all 11 FISH-positive cases in which next-generation sequencing studies could be successfully performed, whereas no BRAF fusions were identified in any of the FISH-negative cases investigated.
We also looked into FISH signal patterns in each positive case and the possible correlation with different fusion partners. Interestingly, all six cases except one with the fusion partner other than SND1 showed the ‘single 3′BRAF’ FISH signal pattern; in contrast, the ‘broken apart’ signal was slightly dominant (4/6) in SND1–BRAF fusions. FISH signal pattern and BRAF fusion partner are listed case by case in Table 1.
Immunohistochemical Staining for pERK
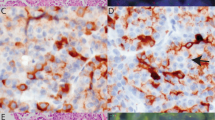
Immunohistochemical staining for pERK, a readout of MAPK pathway activation, was performed on all samples in the discovery cohort and on 11 of 18 cases in the second cohort for which sufficient material was available (Supplementary Table 1). Representative immunohistochemical staining for pERK is shown in Figure 3. Of the fusion-positive cases (n=12), 11 had sufficient material for immunohistochemical analyses; 10 stained strongly positive for pERK and the remaining case showed focal positivity.
Representative immunohistochemical staining for pERK and corresponding morphology feature and BRAF rearrangement FISH analysis finding. (a) A pure acinar cell carcinoma case with solid growth pattern, expressing nuclear and cytoplasmic pERK labeling. BRAF rearrangement was detected by the FISH assay showing multiple ‘single orange (3′BRAF)’ signals; (b) a mixed acinar ductal carcinoma case, positive for pERK staining. BRAF rearrangement was detected by the FISH assay showing ‘broken apart’ signal pattern; (c) a pure acinar cell carcinoma case with glandular growth pattern is focally positive for pERK by immunohistochemistry. BRAF rearrangement was not detected by the FISH assay, but the FISH assay revealed BRAF gain/amplification.
Of the 31 fusion-negative samples with material available for immunohistochemistry, 3 were positive (all in the discovery cohort), 4 were focally positive (1 in the discovery and 3 in the second cohort), and 24 were negative for pERK staining. Cases from the discovery cohort were discussed, regarding the mechanisms for positive pERK staining, in our previous publication.3 In terms of the second cohort, amplification of BRAF and FGFR1 were likely responsible for the activation in the MAPK pathway and pERK focal staining in two samples, respectively. The mechanism for focal pERK staining in the remaining one sample could not be elucidated as next-generation sequencing mutation profiling was not performed due to the insufficiency of tumor material available for the study.
Comparison of BRAF Fusion Status to Clinicopathologic Features
Tumors with BRAF fusion found to be significantly smaller than those without BRAF fusion (median tumor size 3.5 vs 5.5 cm, P=0.001). However, there was no significant difference in other clinicopathologic features, including age, gender, lymphovascular invasion, tumor stage (pT), and lymph node stage (pN). Similarly, BRAF fusions did not bear any prognostic significance (3- and 5-year disease-free survival of the patients whose tumors revealed BRAF fusions were 28 and 14 vs those of the patients whose tumors did not reveal BRAF fusions were 54% and 40%, respectively; P=0.24) (Supplementary Table 3).
Discussion
The discovery of recurrent BRAF fusions in acinar-type neoplasms, including pure acinar cell carcinomas and mixed acinar carcinomas, indicates a new potential therapeutic target for patients whose tumors harbor these changes. In this study, we evaluated the utility of a BRAF-rearrangement FISH assay for the detection of BRAF fusions in acinar-type neoplasms and demonstrated 100% analytical sensitivity and specificity. In BRAF-rearranged cases, the fusion signal being observed in over 60% of the cells analyzed (Avg=84%). Our FISH study also indicated that BRAF gene rearrangements are diffusely present in BRAF-fusion acinar-type neoplasm samples, which is consistent with an oncogenic driver alteration pattern. The relative proximity of SND1 (the most frequent fusion partner involved in BRAF-fusion acinar-type neoplasms) and BRAF at 7q was not an issue for the detection of SND1–BRAF fusions using this test. The ‘single 3′BRAF’ signal pattern (loss of 5′BRAF) was commonly observed in BRAF-rearranged cases (50%), which is compatible with functional studies of previously identified BRAF fusions showing fusions with BRAF truncations of the N terminus function as constitutive kinases through RAF dimerization.3, 8, 9, 10, 11, 12, 13, 14
Compared to the FISH assay, targeted next-generation sequencing-based DNA-seq studies are capable of identifying fusions and also the specific fusion partners and fusion points, provided that the fusions occur within exons or introns included in the assay.5 However, the sensitivity of BRAF fusion detection varies due to the nature of the genomic locus around the fusion point as well as the technical limitations of the methodology. For instance, HERPUD1–BRAF fusions occurred at BRAF exon 7 in cases #1_8 and #2_3 were not detected by MSK-IMPACT because our design does not include capture baits targeting intron 6 of BRAF. Given that multiple BRAF fusion partners occur in acinar-type neoplasms, it is not feasible to design individual RT-PCR tests for diagnostic use. Complementarily, RNA-seq is an ideal method for the detection of fusion transcripts. However, the quality of RNA extracted from formalin-fixed paraffin-embedded tissues may limit the utility of this technical platform. In fact, a few distinct features of FISH, such as fast turn-around time, extremely low sample consumption and tumor content requirement, cost-efficiency, as well as high sensitivity and specificity, make FISH superior to targeted DNA-seq or RNA-seq when a test for a specific fusion is needed in the diagnostic setting.
Prior to the discovery of BRAF fusions in pancreatic acinar-type neoplasms, BRAF fusions had been identified in several other cancer types,8, 9, 10, 11, 13, 14, 15 with an enrichment in pilocytic astrocytomas.9, 14 KIAA1549-BRAF is the most frequent BRAF fusion in pilocytic astrocytomas, followed by FAM131B-BRAF. Interestingly, both fusions are derived from subtle unbalanced genomic alterations at chromosome locus 7q34, a ~2 Mb tamdem duplication and a ~2.5 Mb interstitial deletion, respectively. In contrast, the BRAF fusion partners identified in acinar carcinomas are physically distant from BRAF, with SND1, which is ~13 Mb away from BRAF on chromosome 7, being the closest one. Our study in acinar-type neoplasms demonstrated that the BRAF rearrangement FISH test presented here is fully capable of detecting SND1–BRAF fusions. However, this FISH assay may not be optimal for the detection of KIAA1549-BRAF fusions in pilocytic astrocytomas where BRAF fusions resulted from subtle genomic alterations. In fact, we have developed another BRAF FISH assay that is optimized to screening for KIAA1549-BRAF in pilocytic astrocytomas (data not shown).
As shown in Table 1, BRAF fusions were observed in 11 primary pancreatic samples and one primary hepatic sample, which indicates that hepatic acinar carcinomas can share the same genetic changes as their pancreatic counterparts. The occurrence of acinar-type neoplasm as a primary liver neoplasm has been reported in a few cases.16, 17, 18 In this study, we included four primary liver carcinomas that showed acinar cell differentiation without any evidence of hepatocellular differentiation. For each case, a pancreatic primary was excluded on preoperative imaging procedures, during surgery, and based on follow-up after surgery, thus justifying their classification as primary acinar-type neoplasms of the liver.
In terms of histopathologic subtypes, BRAF gene rearrangements were observed in all variants of acinar-type neoplasms: five pure acinar cell carcinomas, four mixed acinar ductal carcinomas, two mixed acinar neuroendocrine carcinomas, and one acinar neuroendocrine ductal carcinoma. Table 2 illustrates the prevalence of BRAF fusions in each histologic subtype. Although the numbers of cases are relatively small, it is worth noting that 4/5 (80%) mixed acinar ductal carcinomas in our study had BRAF fusions, and all were SND1–BRAF fusions. In contrast, BRAF fusions were much less prevalent in other histologic subtypes, ie, roughly 20% of each of the other subtypes had BRAF fusions. In addition, fusion partners other than SND1 seemed to be predominate in other histologic subtypes, and both HERPUD1–BRAF cases were pure acinar cell carcinomas. Why BRAF fusions, in particular SND1–BRAF, are enriched in mixed acinar ductal carcinomas is unclear; the prevalence of BRAF fusion in pancreatic ductal adenocarcinomas is extremely rare (<0.1%),15 so it seems unlikely that the components with a ductal lineage contributed to the observed increased frequency. Whether or not 5′ partners in BRAF fusions impact clinical outcomes or sensitivity to targeted therapy will be addressed in future studies.
In summary, we developed a BRAF-rearrangement FISH assay to screen for BRAF fusions in pancreatic acinar-type neoplasms. In our expanded study cohort, we demonstrated BRAF fusions in 24% (12/49) of acinar-type neoplasms, a rate similar to that reported in our previous study. In comparison to other sequencing-based technologies, we demonstrated that the FISH assay is highly sensitive and specific as well as time- and cost-efficient in the detection of BRAF fusions in acinar-type neoplasms. The FISH assay can be easily implemented in diagnostic settings to identify acinar-type neoplasm patients suitable for targeted therapy to inhibit MAPK pathway activity.
References
Hruban RH, Pitman MB, Klimstra DS . Acinar neoplasms. In: Silverberg SG (ed). AFIP Atlas of Tumor Pathology: Tumors of the Pancreas, 4th edn. American Registry of Pathology: Washington, DC, USA, 2007, pp 191–214.
Klimstra DS, Adsay V . Acinar neoplasms of the pancreas-A summary of 25 years of research. Semin Diagn Pathol 2016;33:307–318.
Chmielecki J, Hutchinson KE, Frampton GM et al. Comprehensive genomic profiling of pancreatic acinar cell carcinomas identifies recurrent RAF fusions and frequent inactivation of DNA repair genes. Cancer Discov 2014;4:1398–1405.
Wood LD, Klimstra DS . Pathology and genetics of pancreatic neoplasms with acinar differentiation. Semin Diagn Pathol 2014;31:491–497.
Cheng DT, Mitchell TN, Zehir A et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–264.
Zheng Z, Liebers M, Zhelyazkova B et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479–1484.
Furukawa T, Sakamoto H, Takeuchi S et al. Whole exome sequencing reveals recurrent mutations in BRCA2 and FAT genes in acinar cell carcinomas of the pancreas. Sci Rep 2015;5:8829.
Ciampi R, Knauf JA, Kerler R et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest 2005;115:94–101.
Jones DT, Kocialkowski S, Liu L et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 2008;68:8673–8677.
Palanisamy N, Ateeq B, Kalyana-Sundaram S et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 2010;16:793–798.
Lee NV, Lira ME, Pavlicek A et al. A novel SND1-BRAF fusion confers resistance to c-Met inhibitor PF-04217903 in GTL16 cells through [corrected] MAPK activation. PLoS ONE 2012;7:e39653.
Sievert AJ, Lang SS, Boucher KL et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci USA 2013;110:5957–5962.
Hutchinson KE, Lipson D, Stephens PJ et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res 2013;19:6696–6702.
Cin H, Meyer C, Herr R et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol 2011;121:763–774.
Ross JS, Wang K, Chmielecki J et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138:881–890.
Wildgruber M, Rummeny EJ, Gaa J . Primary acinar cell carcinoma of the liver. RoFo 2013;185:572–573.
Zundler S, Erber R, Agaimy A et al. Pancreatic panniculitis in a patient with pancreatic-type acinar cell carcinoma of the liver—case report and review of literature. BMC Cancer 2016;16:130.
Agaimy A, KaiserA, Becker K et al. Pancreatic-type acinarcell carcinoma of the liver: a clinicopathologic study of four patients. Mod Pathol 2011;24:1620–1626.
Acknowledgements
Research reported in this publication was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Juliann Chmielecki, Jeffrey Ross, and Philip J Stephens are employees and stockholders in Foundation Medicine.
Additional information
This study was presented in part at the 103rd annual meeting of the United States and Canadian Academy of Pathology in Boston, MA, in March 2015.
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Wang, L., Basturk, O., Wang, J. et al. A FISH assay efficiently screens for BRAF gene rearrangements in pancreatic acinar-type neoplasms. Mod Pathol 31, 132–140 (2018). https://doi.org/10.1038/modpathol.2017.106
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.106
This article is cited by
-
RET gene rearrangements occur in a subset of pancreatic acinar cell carcinomas
Modern Pathology (2020)
-
RAF1 rearrangements are common in pancreatic acinar cell carcinomas
Modern Pathology (2020)