Abstract
Müllerian adenosarcomas are malignant gynecologic neoplasms. Advanced staging and sarcomatous overgrowth predict poor prognosis. Because the genomic landscape remains poorly understood, we conducted this study to characterize the genomewide copy number variations in adenosarcomas. Sixteen tumors, including eight with and eight without sarcomatous overgrowth, were subjected to a molecular inversion probe array analysis. Copy number variations, particularly losses, were significantly higher in cases with sarcomatous overgrowth. Frequent gains of chromosomal 12q were noted, often involving cancer-associated genes CDK4 (six cases), MDM2, CPM, YEATS4, DDIT3, GLI1 (five each), HMGA2 and STAT6 (four), without association with sarcomatous overgrowth status. The most frequent losses involved chromosomes 13q (five cases), 9p, 16q and 17q (four cases each) and were almost limited to cases with sarcomatous overgrowth. MDM2 and CDK4 amplification, as well as losses of RB1 (observed in two cases) and CDKN2A/B (one case), was verified by FISH. By immunohistochemistry, all MDM2/CDK4-coamplified cases were confirmed to overexpress both encoded proteins, whereas all four cases with (plus an additional four without) gain of HMGA2 overexpressed the HMGA2 protein. Both cases with RB1 loss were negative for the immunostaining of the encoded protein. Chromothripsis-like copy number profiles involving chromosome 12 or 14 were observed in three fatal cases, all of which harbored sarcomatous overgrowth. With whole chromosome painting and deconvolution fluorescent microscopy, dividing tumor cells in all three cases were shown to have scattered extrachromosomal materials derived from chromosomes involved by chromothripsis, suggesting that this phenomenon may serve as visual evidence for chromothripsis in paraffin tissue. In conclusion, we identified frequent chromosome 12q amplifications, including loci containing potential pharmacological targets. Global chromosomal instability and chromothripsis were more frequent in cases with sarcomatous overgrowth. To our knowledge, this is the first time that evidence of chromothripsis has been demonstrated in paraffin-embedded clinical tissues and in adenosarcomas.
Similar content being viewed by others
Main
Müllerian adenosarcomas are malignant mixed epithelial–mesenchymal neoplasms that arise most frequently in the uterine corpus.1, 2 They account for approximately 5% of all primary uterine sarcomas and can affect women from a wide age range.3, 4 Most Müllerian adenosarcomas present as polypoid endometrial masses, and can be accompanied by extensive myometrial invasion and extrauterine spread. Histologically, adenosarcomas display a biphasic appearance with admixed sarcoma components and benign-appearing Müllerian glands. Sarcomatous overgrowth, defined as the presence of at least 25% tumor area overgrown by a pure sarcomatous component, is seen in a subset of cases.2, 5 Clinically, a majority of adenosarcomas present with low-stage disease and follow an indolent clinical course after surgical treatment. Tumors showing sarcomatous overgrowth or deep myometrial invasion, however, behave more aggressively.4, 6, 7, 8 Adenosarcomas less frequently occur in the ovary, where they pose a greater risk of peritoneal spread and a worse prognosis.8 The mainstay of treatment for localized Müllerian adenosarcomas is surgery,9 whereas more advanced diseases often entail other modalities of treatment, although the roles of adjuvant chemotherapy and/or radiation therapy in the management of adenosarcomas are not well-defined. The previous lack of knowledge regarding the molecular and genetic basis of Müllerian adenosarcomas has hindered our ability to develop tumor-specific therapeutic strategies.
Recently, the unraveling of the genetic background of adenosarcomas has begun. A panel of 275 cancer-associated genes were interrogated on adenosarcomas from 18 patients with targeted sequencing, and the most frequent findings included MDM2 and CDK4 amplification (28%) as well as mutations involving a variety of PIK3CA/AKT/PTEN pathway members (72%).10 A more recent study employing whole-exome, target-capture and RNA sequencing found amplification of MDM2/CDK4/HMGA2 and TERT genes, recurrent mutations of FGFR2, KMT2C, and DICER1 genes, as well as nuclear receptor coactivator family gene rearrangement, each in a minority of cases.11 However, an overview of the chromosomal aberrations at a genomewide level is still lacking. In this report, we employed a molecular inversion probe array to characterize the genomewide copy number variations in 16 adenosarcomas, with an emphasis on the genomic landscape that defined sarcomatous overgrowth.
Materials and methods
Material Collection and Pathological Review
Primary Müllerian adenosarcomas diagnosed between 1995 and 2014 were identified from the pathology archives of National Taiwan University Hospital (Taipei, Taiwan). The histology was reviewed by three pathologists (JCL, K-TK, and M-CL). The diagnosis of adenosarcoma and sarcomatous overgrowth was based on the criteria adopted by the World Health Organization.2 All available cases with sarcomatous overgrowth were included, and the same number of cases without sarcomatous overgrowth was selected according to the availability of adequate tumor proportion (≧75%; cases with the highest tumor proportion were preferred). This study was approved by the National Taiwan University Hospital Research Ethics Committee.
Molecular Inversion Probe Array Analysis
At least 80 ng genomic DNA extracted from each sample was subjected to the molecular inversion probe array using the OncoScan FFPE Assay Kit (Affymetrix, Santa Clara, CA) as previously described.12 Arbitrary cutoffs were used to define a homozygous deletion, a loss, and a gain with copy numbers of <0.5, <1.5, and >3, respectively, as previously described.13 To evaluate the quantitative level of genomewide copy number changes, a chromosome instability index was calculated as previously described.13 As a modification, the length of the copy number variation segments was also taken into consideration by multiplying the length of each segment by its inferred copy number. Subsequently, Wilcoxon rank-sum test was performed to assess the significance level of the chromosome instability values between different groups. The cutoff significance level was 0.05. The microarray data have been submitted to the Gene Expression Omnibus with accession number GSE67107 (http://www.ncbi.nlm.nih.gov/pubmed/11752295).
Fluorescence In Situ Hybridization
Four-μm-thick tissue slides were used for fluorescence in situ hybridization (FISH) analysis. The FISH probes used included Vysis LSI MDM2/CEP12 probe and Vysis CDKN2A/CEP9 probe (also covering the CDKN2B locus; Abbott Molecular, Des Plaines, IL); CDK4/SE12 probe (Leica Biosystems, Nußloch, Germany); ZytoLight SPEC RB1/13q12 dual color probe (ZytoVision GmbH, Bremerhaven, Germany); and whole chromosome 12 and 14 painting probes (Applied Spectral Imaging, Carlsbad, CA) The results were observed under a fluorescent microscope (Axio Imager 2, Carl Zeiss MicroImaging GmbH, Jena, Germany) by a pathologist (CWL) blinded to the clinical and molecular data. For MDM2 and CDK4 copy numbers, 20 nuclei were counted for each case; the consensus criteria recommended for HER2 FISH were adopted for interpretation of the results.14 For CDKN2A/B and RB1 copy numbers, 50 nuclei were counted; monoallelic deletion was defined as >50% nuclei harboring only 1 signal of respective locus, and relative deletion was defined as a CDKN2A/CEP9 or RB1/13q12 ratio of <0.8 in the absence of monoallelic deletion, as previously described.15
For chromosome painting, the dividing tumor cells at M-phase were searched for. Image acquisition was performed with wide-field DeltaVision deconvolution microscope (Applied Precision, South El Monte, CA), as previously described,16 by an experienced technician (CAC) blinded to the molecular inversion probe array data. Stacks of optical section images were collected through SoftWorX application suite software. Both 2D and 3D images were generated and analyzed with VoloCity software (Perkin-Elmer, Waltham, MA).
Immunohistochemistry
Four-μm-thick tissue slides were subjected to immunohistochemistry using the following primary antibodies: MDM2 (Invitrogen, Grand Island, NY; clone 1F2; dilution 1:50), CDK4 (Santa Cruz, Santa Cruz, CA; clone C-22; dilution 1:800), HMGA2 (BioCheck, Foster City, CA; dilution 1:2000), and retinoblastoma gene protein (RB; Leica Biosystems, Nußloch, Germany; clone 1F8; dilution 1:100). A complete loss of nuclear RB expression in the tumor cells was considered as a negative result. For the remaining immunostains, either a diffuse (>50% tumor cells) and at least moderate or focal (10–50%) but strong nuclear staining pattern (obscuring nuclear contours) was considered as a positive result.
The reader is referred to the Online Methods in Supplementary Information for more details about the molecular inversion probe array analysis results, FISH, and immunohistochemistry.
Results
Clinicopathologic Features of the Study Groups
Fifteen uterine and one ovarian Müllerian adenosarcomas were submitted for molecular inversion probe array analysis, including all eight cases with sarcomatous overgrowth (ie, cases S1–8 where ‘S’ stands for Sarcomatous overgrowth) as well as eight without (ie, cases N1–8 where ‘N’ stands for Non-sarcomatous overgrowth). Four other available cases without sarcomatous overgrowth were excluded because of relatively low neoplastic cell purity even with tissue coring. The patients’ ages ranged from 22 to 77 years (median 50 years). During a median follow-up of 11 months, seven tumors with sarcomatous overgrowth claimed the respective patients’ lives; by contrast, all patients in the ‘N’ group were alive without evidence of disease after a median follow-up of 47.5 months. Table 1 presents the clinicopathologic features.
Genomewide Copy Number Variations
Frequent (appearing in at least 25% or four cases) gains were limited to various segments on chromosome 12 (p12.3 and q13.3–21.13). Gains of 12q13.3-q14.1 (encompassing CDK4) were noted in six cases (37.5%). In five, five, and four cases, respectively, the amplicon that contained CDK4 also included DDIT3, GLI1, and STAT6 loci. Gains of 12q15 (including CPM, MDM2, and YEATS4) were observed in five cases. Gains of the HMGA2 gene at 12q14.3 were found in four cases, all of which also had concurrent gains of the aforementioned genes in chromosome 12q13.3-q14.1. Gains of the entire or partial PIK3C2G gene on 12p12.3 were noted in five cases. Otherwise, frequent gains were also found in 5p, 7p, 8q, and 14q.
On the other hand, frequent losses were observed in chromosomes 9p, 13q, 15q, 16q, 17q, and 22q. Except for 15q13.3 and 22q11.23 (containing GSTT1 gene locus, which might represent a germline polymorphism17), most of these chromosomal losses were predominantly observed in the ‘S’ group of cases. Homozygous deletions were infrequent. Figure 1 illustrates the overall and group-wise prevalence of gains and losses across all chromosomes. Supplementary Figure S1 demonstrates the genomewide copy number variations of each case. Table 2 lists chromosomal regions with frequent gains or losses and the genes encompassed.
Validation of Copy Number Changes by FISH and Immunohistochemistry
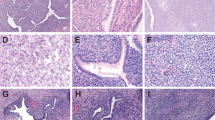
As MDM2 and CDK4 gains were the most frequent copy number variations in the current cohort and well-established commercial FISH probes and antibodies were available for MDM2 and CDK4, we chose these two genes for further analysis to validate the status of gain derived from the molecular inversion probe array data. As a result, coamplification of both genes was confirmed in all five cases by FISH, with concomitant overexpression of MDM2 and CDK4 proteins as demonstrated with immunohistochemistry. In cases belonging to the ‘S’ group, the sarcomatous-overgrowth and non-sarcomatous-overgrowth components in each tumor showed similar copy number status. Figure 2a–e demonstrates the histology, FISH, and immunostains of a representative case. The glandular components of four adenosarcomas that had MDM2/CDK4 coamplification was included in the sections for FISH analysis, and none of the admixed glands showed amplification of either gene (Figure 2b and c and Supplementary Figure S2A). By contrast, none of the remaining 11 tumors had amplification of either gene or overexpressed respective encoded proteins (Figure 2f–j) except for case N7, which exhibited focal positive CDK4 immunostaining. It is noteworthy that case S2, where low-level gain of CDK4 gene (with inferred copy number of 3.39) was revealed by microarray, showed chromosome 12 polysomy by FISH (with 3.35 and 4.00 copies of centromere and CDK4, respectively) and negative CDK4 immunostaining.
Histology, MDM2 FISH, CDK4 FISH, MDM2 immunostaining, and CDK4 immunostaining in cases with (a–e, case N1) and without (f–j, case N3) MDM2/CDK4 coamplification. For MDM2 and CDK4 FISH, red signals indicate MDM2 or CDK4 loci, while green signals indicate centromeres of chromosome 12. Note that the benign glandular epithelia (delineated by yellow-dashed lines in the left-lower corner of b and c) do not harbor amplification of either gene.
To verify the losses detected by molecular inversion probe array while considering the potential biological relevance and availability of reliable commercial probes, the copy numbers of tumor suppressors CDKN2A/B (9p21.3) and RB1 (13q14.2) were further validated in 12 cases (including all cases with inferred loss) by FISH. Interpretable results were obtained in 11 and 12 cases, respectively. Consistent with the microarray data, monoallelic RB1 deletions were corroborated in cases S1 and S6. Furthermore, both cases proved to be negative for RB protein by immunohistochemistry. Of the three cases with inferred CDKN2A/B loss, monoallelic deletion was confirmed by FISH in S6, whereas the other two cases demonstrated relative deletion of this locus with chromosome 9 polysomy. Meanwhile, the remaining cases analyzed showed no evidence of deletion of either locus. In cases where deletions were confirmed in the sarcomatous-overgrowth component, those deletions were also conspicuously present in the matched non-sarcomatous-overgrowth areas with similar copy numbers. Representative FISH and immunohistochemistry results are shown in Supplementary Figure S2B–F and Supplementary Table S1.
Previous studies reported HMGA2 amplification or overexpression, respectively, without matched copy number and protein expression data addressing the possible causative relationship.10, 11 To correlate the protein expression status with our molecular inversion probe array findings, HMGA2 immunostaining was performed in this series and was positive in all four cases with HMGA2 gains (Supplementary Figure S2G–I). It is noteworthy that four additional cases without HMGA2 gain also showed positive immunostaining, indicating other mechanisms of upregulating HMGA2 expression. Taken together, these analyses validated the capability of the current microarray-based platform to detect gains and losses using the current cutoff thresholds.
Chromosome Instability Assessment and Clinicopathologic Correlation
To determine whether the phenomenon of sarcomatous overgrowth might reflect genomewide chromosomal instability, the chromosome instability indices were compared between the two groups (Figure 3). As expected, the ‘S’ group had significantly higher chromosome instability indices than the ‘N’ group (mean 198.3 vs 41.9; P=0.0002). When considered separately, both losses and gains contributed to the higher chromosomal instability of the ‘S’ group (mean 124.8 vs 9.1 for losses and 117.7 vs 32.8 for gains), although the statistical significance was substantially higher for the part of losses by a difference of two orders of magnitude (P=0.0002 for losses vs P=0.0165 for gains; Figure 3). By contrast, as the most frequently amplified loci, the copy number status of MDM2 and CDK4 was not significantly associated with the chromosome instability index (mean 147.8 vs 107.6 in cases with and without MDM2/CDK4 coamplification; P=0.3773). The only surviving case in the ‘S’ group (S7) had the lowest chromosome instability index attributed to chromosomal losses (but not gains) among this group.
From left to right: Overall chromosome instability indices as well as the fractions contributed by gains and losses, respectively, of the two groups of cases. On the right: Overall chromosome instability indices of cases with vs without MDM2/CDK4 coamplification. (Error bar: mean±95% confidence interval. Asterisks denote reaching statistical significance, with their number indicating the difference in the order of magnitude.)
Identification of Chromothripsis-Like Copy Number Profiles with Chromosome Painting Correlation
In three clinically fatal tumors harboring sarcomatous overgrowth, conspicuous fluctuation of copy number with neighboring chromosomal regions showing oscillations toward different directions (eg, gain vs loss) was noted in chromosome 12q13.11–q15 (S1), 12q13.12-q21.31 (S6), and 14q21.1–q24.2 (S5), respectively (Figure 4). This highly fragmented status involving single chromosomes indicated the phenomenon of chromothripsis.18, 19 Both cases S1 and S6 also harbored amplifications involving chromosome 12 (including CDK4 and MDM2 genes), implying that chromothripsis might be the mechanism accounting for amplification of these loci. In comparison, cases N1, N2, and S3 also had amplifications of these loci despite no evidence of chromothripsis (Figure 4), thus implying a different mechanism of amplification.
Analyses of molecular inversion probe array data demonstrate chromothripsis-like pattern of copy number oscillation in chromosome 12 in case S6 (a, b) and case S1 (c), as well as chromosome 14 in case S5 (d). For comparison, case N1 shows chromosome 12 amplification without evidence of chromothripsis (e).
To further visualize the status of respective chromosomes involved by chromothripsis, the three cases were then subjected to chromosome 12 and 14 painting (Figure 5a and b, Supplementary Figure S3A, and Supplementary Videos 1 and 2). Long intact segments of chromosome 14 were observed in cases S1 and S6, which was also true for chromosome 12 in case S5. By contrast, small scattered fragments, in addition to normal-appearing long segments, composed of chromosome 12 material were observed in cases S1 and S6; this was also true for chromosome 14 in case S5. This was consistent with chromothripsis-like profiles of respective chromosomes shown by the molecular inversion probe array data. Case N1 served as the control and showed intact chromosome 12 segments despite MDM2 amplification, again indicating a distinct mechanism of MDM2 amplification in lack of chromosome 12 chromothripsis (Supplementary Figure S3B). Furthermore, mixed chromosome 12 painting and MDM2/CEP12 probes were applied to cases S1 and N1 to compare the patterns of MDM2 amplification in cases with vs without chromosome 12 chromothripsis. Interestingly, in case S1 the amplified MDM2 signals were scattered mainly in extrachromosomal locations, whereas those in case N1 tended to be located on an elongated arm of chromosome 12 (Figure 5c and d and Supplementary Videos 3 and 4).
Chromosome 12 and 14 dual painting in case S1 (a) and case S5 (b). Note that chromosome 14 (labeled in red) in (a) and chromosome 12 (green) in (b) are mainly presented as long intact segments, whereas scattered tiny extrachromosomal materials derived from chromosome 12 (a) and chromosome 14 (b) are additionally observed. (c, case S1 and d, case N1) Mixed chromosome 12 painting and MDM2 FISH. Red signals indicate MDM2 loci and strong green signals indicate centromeres of chromosome 12 (CEP12 probe). Chromosome 12 is painted in weak green and artificially outlined by yellow-dashed lines for visual clarity. Note that in case S1 the amplified MDM2 signals are largely scattered outside the parental chromosome 12, whereas in case N1 the amplified MDM2 signals are distributed along an elongated arm of chromosome 12. See also Supplementary Videos 1, 2, 3, and 4 for three-dimensional reconstruction of (a), (b), (c), and (d), respectively.
Discussion
The genetic basis of Müllerian adenosarcomas had been poorly defined until recent efforts started to shed light on this rare neoplasm. By employing next-generation sequencing-based platforms, amplifications of MDM2, CDK4, HMGA2, MYBL1, TERT genes, mutations of PIK3CA/AKT/PTEN pathway members and FGFR2, KMT2C and DICER1 genes, as well as rearrangement of nuclear receptor coactivator family members were each disclosed in a subset of cases.10, 11 However, the genomic copy number landscape, especially the one that correlates with sarcomatous overgrowth and aggressive biology, has not been fully explored. In the present study, we were able to characterize the genomewide copy number variation in 16 adenosarcomas for the first time. In line with the previous report,10, 11 concurrent gains of loci in 12q13–15 containing CDK4 and MDM2 were the most frequent copy number variation events. Known as oncogenes involved in cell-cycle regulation and progression, MDM2 and/or CDK4 has been implicated in a wide spectrum of human cancers.20, 21, 22, 23, 24, 25 In dedifferentiated liposarcoma, for instance, amplification of MDM2 is nearly always accompanied by CPM, HMGA2, and YEATS4 in the same amplicon, while CDK4 can sometimes have GLI1 and DDIT3 as the amplification partners.26, 27 These findings are similar to what has been observed in our adenosarcoma cases. Interestingly, the amplification of MDM2 and CDK4 seemed to be limited to the mesenchymal component, suggesting a non-neoplastic nature of the glandular component, consistent with the recent findings.11 Finally, selective therapeutic reagents antagonizing MDM2 and CDK4 have shown some efficacy in clinical trials in other tumor types with amplifications of respective genes (http://www.clinicaltrials.gov/)28, 29 Therefore, patients with advanced stage adenosarcomas should be tested for MDM2 and CDK4 amplification and allowed access to these emerging therapies.
HMGA2 is a chromatin-associated protein and transcriptional regulator. Rearrangement and resultant overexpression of HMGA2 has been implicated in a variety of human neoplasms.30 Whereas our data confirmed that all four cases with gains of HMGA2 also expressed the encoded protein, four additional cases without the gain expressed HMGA2 protein as well, indicating alternative mechanisms of upregulating this protein, such as genetic rearrangement. Interestingly, HMGA2 rearrangement or amplification has been implicated in some endometrial polyps.31, 32 Therefore, the finding of frequent HMGA2 amplification and overexpression in adenosarcoma perhaps provides a possible tumorigenic link between both of these benign and malignant counterparts of polypoid endometrial mesenchymal tumors.
Whereas gains of chromosome 12q seemed evenly distributed to tumors regardless of the status of sarcomatous overgrowth, chromosomal losses were obviously enriched in more aggressive tumors with sarcomatous overgrowth. Among the most frequent losses disclosed in this study, those involving chromosomal segments in 9p, 13q, 16q, and 17q were almost limited to the ‘S’ group of tumors. Some well-known tumor suppressors were identified in the recurrently deleted loci, including NF1 (17q11.2), CDKN2A and CDKN2B (9p21), RB1 (13q14), as well as TP53 (17p13.1), which were deleted in four, three, two, and two cases, respectively, all of them belonging to the ‘S’ group. In addition, a parallel comparative FISH analysis confirmed that the losses of CDKN2A/B and RB1 were present in both the sarcomatous overgrowth and non-overgrowth areas of cases analyzed. Collectively, these observations suggested that the partial loss of these tumor suppressors preceded, and possibly set the stage for, the full-blown sarcomatous overgrowth.
Global chromosomal instability, as quantified with a chromosome instability index, was clearly associated with more aggressive tumors in the present study, a phenomenon that has been observed in other human cancers including gynecological malignancies.33, 34 Notably, the amplifications of loci containing MDM2 and CDK4 did not significantly correlate with the chromosome instability index, suggesting that these changes might not be randomly secondary to global chromosomal instability. Together with its frequent occurrence, this finding indicated critical roles for MDM2 and CDK4 amplification in the tumorigenesis of a subset of adenosarcomas. By contrast, chromosomal losses preferentially occurred in the ‘S’ group of tumors, which had significantly higher chromosome instability indices than the ‘N’ group. This suggests that some elements in the frequently deleted regions could have critical roles in maintaining chromosomal integrity, possibly including some of the aforementioned tumor suppressors that have been implicated in chromosomal instability.35, 36
Chromothripsis has been described as a cellular catastrophe that causes pulverization of one or a few chromosomes, typically with massive rearrangements of the affected chromosome regions accompanied by numerous copy number losses and gains.18, 37 To infer chromothripsis with array-based copy number profiling data, various criteria have been applied, the central common part of which is to identify oscillating copy number states on a single chromosome.38, 39, 40, 41 On the basis of copy number profiling, chromothripsis was present in an estimated 1–3% of human cancers.18, 42 By contrast, 19% (3/16) of cases (and 38% of the ‘S’ group) in the current series presented copy number profiles suggesting chromothripsis, a phenomenon unprecedented in adenosarcoma. All the three tumors belonged to the ‘S’ group, which is consistent with previous observations that chromothripsis is generally correlated with poor prognosis.19, 43 It is worth noting that this series was enriched for tumors with sarcomatous overgrowth with the intent to compare and disclose key features associated with aggressiveness, thus leading to a possible overestimation of the overall incidence of chromothripsis in adenosarcoma. Furthermore, by whole chromosome painting, we observed small extrachromosomal fragments derived from chromosomes that had chromothripsis-like copy number profiles. It has been implied that chromothripsis causes amplifications in the form of double minutes.18, 38 The chromosome painting analysis suggested that amplification of chromosome 12 in cases S1 and S6 might have resulted from chromothripsis through double minute formation, whereas amplifications in chromosomes without chromothripsis might have been caused by other mechanisms.44 Indeed, the mixed chromosome 12 painting and MDM2 FISH in Figure 5d might give a hint of homogeneously staining region as the mechanism of amplification.
Collectively, these results imply the possible utility of chromosomal painting, in conjunction with high-resolution microscopy, as a surrogate for high-throughput platforms to detect chromothripsis. Importantly, this approach is also applicable to FFPE samples, which are generally deemed suboptimal for genomic research. However, it does have significant limitations. First, chromothripsis unaccompanied by conspicuous amplification should evade detection. Second, this approach seems pragmatic only in tumors with frequent amplification of certain well-established loci (such as those containing MDM2 and CDK4 in liposarcomas). Third, as chromosome painting aims at dividing cells to obtain interpretable results, it may not be suitable for tumors with low mitotic count. Finally, it remains to be determined whether the current evidence of chromothripsis (as detected by either microarray or chromosome painting) represents true chromothripsis or not. Next-generation sequencing-based methods may be required for validation.
In conclusion, we characterized the genomewide copy number variation in 16 Müllerian adenosarcomas and observed frequent amplification of chromosome 12q13-q21 containing oncogenes such as MDM2, YEATS4, CDK4, and HMGA2, some being potential therapeutic targets. Global chromosomal instability, particularly in association with increased copy number losses and chromothripsis-like features, was frequently found in clinically aggressive tumors with sarcomatous overgrowth. Finally, chromothripsis-like copy number profiles corresponded to a distinct FISH pattern that implied extrachromosomal double minute formation.
References
Clement PB, Scully RE . Mullerian adenosarcoma of the uterus. A clinicopathologic analysis of ten cases of a distinctive type of mullerian mixed tumor. Cancer 1974;34:1138–1149.
Wells MOE, Palacios J, Prat J Mixed epithelial and mesenchymal tumours. In: Kurman R, Carcangiu M, Herrington C et al (eds). WHO Classification of Tumours of Female Reproductive Organs, 4th edn. IARC: Lyon, France, 2014.
Major FJ, Blessing JA, Silverberg SG et al. Prognostic factors in early-stage uterine sarcoma. A Gynecologic Oncology Group study. Cancer 1993;71:1702–1709.
Abeler VM, Royne O, Thoresen S et al. Uterine sarcomas in Norway. A histopathological and prognostic survey of a total population from 1970 to 2000 including 419 patients. Histopathology 2009;54:355–364.
Clement PB . Mullerian adenosarcomas of the uterus with sarcomatous overgrowth. A clinicopathological analysis of 10 cases. Am J Surg Pathol 1989;13:28–38.
Kaku T, Silverberg SG, Major FJ et al. Adenosarcoma of the uterus: a Gynecologic Oncology Group clinicopathologic study of 31 cases. Int J Gynecol Pathol 1992;11:75–88.
Krivak TC, Seidman JD, McBroom JW et al. Uterine adenosarcoma with sarcomatous overgrowth vs uterine carcinosarcoma: comparison of treatment and survival. Gynecol Oncol 2001;83:89–94.
Eichhorn JH, Young RH, Clement PB et al. Mesodermal (mullerian) adenosarcoma of the ovary: a clinicopathologic analysis of 40 cases and a review of the literature. Am J Surg Pathol 2002;26:1243–1258.
Clement PB, Scully RE . Mullerian adenosarcoma of the uterus: a clinicopathologic analysis of 100 cases with a review of the literature. Hum Pathol 1990;21:363–381.
Howitt BE, Sholl LM, Dal Cin P et al. Targeted genomic analysis of Mullerian adenosarcoma. J Pathol 2015;235:37–49.
Piscuoglio S, Burke KA, Ng CK et al. Uterine adenosarcomas are mesenchymal neoplasms. J Pathol 2016;238:381–388.
Wang Y, Cottman M, Schiffman JD . Molecular inversion probes: a novel microarray technology and its application in cancer research. Cancer Genet 2012;205:341–355.
Kuo KT, Guan B, Feng Y et al. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res 2009;69:4036–4042.
Wolff AC, Hammond ME, Hicks DG et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997–4013.
Rajaram V, Leuthardt EC, Singh PK et al. 9p21 and 13q14 dosages in ependymomas. A clinicopathologic study of 101 cases. Mod Pathol 2004;17:9–14.
Changou CA, Chen YR, Xing L et al. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc Natl Acad Sci USA 2014;111:14147–14152.
Pemble S, Schroeder KR, Spencer SR et al. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J 1994;300 (Pt 1):271–276.
Stephens PJ, Greenman CD, Fu B et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011;144:27–40.
Cai H, Kumar N, Bagheri HC et al. Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens. BMC Genomics 2014;15:82.
Wade M, Li YC, Wahl GM . MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013;13:83–96.
Shadfan M, Lopez-Pajares V, Yuan ZM . MDM2 and MDMX: alone and together in regulation of p53. Transl Cancer Res 2012;1:88–89.
Momand J, Zambetti GP, Olson DC et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992;69:1237–1245.
Matsushime H, Ewen ME, Strom DK et al. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 1992;71:323–334.
Baker SJ, Reddy EP . CDK4: a key player in the cell cycle, development, and cancer. Genes Cancer 2012;3:658–669.
Ortega S, Malumbres M, Barbacid M . Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta 2002;1602:73–87.
Italiano A, Bianchini L, Keslair F et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer 2008;122:2233–2241.
Erickson-Johnson MR, Seys AR, Roth CW et al. Carboxypeptidase M: a biomarker for the discrimination of well-differentiated liposarcoma from lipoma. Mod Pathol 2009;22:1541–1547.
Ray-Coquard I, Blay JY, Italiano et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol 2012;13:1133–1140.
Dickson MA, Tap WD, Keohan ML et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol 2013;31:2024–2028.
Cleynen I, Van de Ven WJ . The HMGA proteins: a myriad of functions (Review). Int J Oncol 2008;32:289–305.
Dal Cin P, Wanschura S, Kazmierczak B et al. Amplification and expression of the HMGIC gene in a benign endometrial polyp. Genes Chromosomes Cancer 1998;22:95–99.
Bol S, Wanschura S, Thode B et al. An endometrial polyp with a rearrangement of HMGI-C underlying a complex cytogenetic rearrangement involving chromosomes 2 and 12. Cancer Genet Cytogenet 1996;90:88–90.
Carter SL, Eklund AC, Kohane IS et al. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 2006;38:1043–1048.
Cope L, Wu RC, Shih Ie M et al. High level of chromosomal aberration in ovarian cancer genome correlates with poor clinical outcome. Gynecol Oncol 2013;128:500–505.
Kramer A, Neben K, Ho AD . Centrosome replication, genomic instability and cancer. Leukemia 2002;16:767–775.
Hernando E, Nahle Z, Juan G et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 2004;430:797–802.
Korbel JO, Campbell PJ . Criteria for inference of chromothripsis in cancer genomes. Cell 2013;152:1226–1236.
Rausch T, Jones DT, Zapatka M et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 2012;148:59–71.
Hirsch D, Kemmerling R, Davis S et al. Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer Res 2013;73:1454–1460.
Magrangeas F, Avet-Loiseau H, Munshi NC et al. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood 2011;118:675–678.
Molenaar JJ, Koster J, Zwijnenburg DA et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012;483:589–593.
Kim TM, Xi R, Luquette LJ et al. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res 2013;23:217–227.
Kloosterman WP, Koster J, Molenaar JJ . Prevalence and clinical implications of chromothripsis in cancer genomes. Curr Opin Oncol 2014;26:64–72.
Matsui A, Ihara T, Suda H et al. Gene amplification: mechanisms and involvement in cancer. Biomol Concepts 2013;4:567–582.
Acknowledgements
We thank Professor Sung-Liang Yu, Ms Yuan-Chun Yang (Taipei, Taiwan), Dr Adrián Mariño-Enríquez (Boston, USA), and Dr Jui Lan (Kaohsiung, Taiwan) for their generous help. The microarray data have been submitted to the Gene Expression Omnibus with accession number GSE67107 (http://www.ncbi.nlm.nih.gov/pubmed/11752295). This work was supported by grant NTUH103-S2375 (to K-TK) and NTUH104-S2780 (to M-CL) from National Taiwan University Hospital, Taipei, Taiwan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, JC., Lu, TP., Changou, C. et al. Genomewide copy number analysis of Müllerian adenosarcoma identified chromosomal instability in the aggressive subgroup. Mod Pathol 29, 1070–1082 (2016). https://doi.org/10.1038/modpathol.2016.99
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.99
This article is cited by
-
Atypical uterine polyps show morphologic and molecular overlap with mullerian adenosarcoma but follow a benign clinical course
Modern Pathology (2022)
-
Endometriale und weitere seltene uterine Sarkome
Der Pathologe (2022)
-
Toward a unifying entity that encompasses most, but perhaps not all, inflammatory leiomyosarcomas and histiocyte-rich rhabdomyoblastic tumors
Modern Pathology (2021)
-
Genomic alterations in gynecological malignancies: histotype-associated driver mutations, molecular subtyping schemes, and tumorigenic mechanisms
Journal of Human Genetics (2021)
-
Recurrent MEIS1-NCOA2/1 fusions in a subset of low-grade spindle cell sarcomas frequently involving the genitourinary and gynecologic tracts
Modern Pathology (2021)