Abstract
Extranodal natural killer (NK)/T-cell lymphoma (ENKTL), nasal type, comprises NK or cytotoxic T cells. We evaluated the clinical impact of cell type and the usefulness of T-cell receptor (TCR) gene transcripts in distinguishing cell lineage. One hundred and eight cases of ENKTL were analyzed for TCR gene rearrangements using the BIOMED-2 protocol and for TCR gene expression using immunohistochemistry for TCR-βF1 and TCR-cγM1, and RNA in situ hybridization for TCR gene transcripts. Prognostic factors were analyzed. Among the 108 cases, 44 were monoclonal for a TCR rearrangement (40%) while 64 (60%) were undefinable. The monoclonal cases expressed TCR-βF1 in 14 out of 40 cases (35%) and TCR-cγM1 in 1 out of 44 cases (2%). The 64 undetermined cases expressed TCR-βF1 in 15 cases (23%) and TCR-cγM1 in 1 (2%). Thirteen of 40 TCR-β constant gene transcript-positive cases (33%) expressed TCR-βF1 and one of nine TCR-γ constant gene transcript-positive cases (11%) expressed TCR-cγM1. TCR gene transcripts were not useful in the distinction of cell lineages. TCR gene transcripts were positive in ENKTLs as well as in normal B cells and aggressive NK-cell leukemia. Based on gene rearrangements and immunohistochemistry for TCR, there were 60 T-cell type cases (56%), 32 NK-cell type cases (30%), and 16 cases with an undetermined cell type (14%). TCR protein was expressed in 30/60 T-ENKTLs (50%) in a variable fraction of tumor cells. There were no significant differences in clinical findings or overall patient survival between T- or NK-cell types of ENKTL, although those with a T-cell type tended to show a better prognosis for those with localized nasal lymphomas. Univariate and multivariate analysis showed that a non-nasal ENKTL, age >60 years, high level of lactate dehydrogenase, bone marrow involvement, and the absence of radiotherapy were independent prognostic factors.
Similar content being viewed by others
Main
Extranodal natural killer (NK)/T-cell lymphoma (ENKTL) is a prototype of Epstein–Barr virus (EBV)-positive cytotoxic T- or NK-cell lymphomas predominantly involving the nasal cavity and extranasal upper aerodigestive tract with less frequent involvement of non-nasal sites such as the skin, gastrointestinal tract, soft tissue, and testis.1, 2 The tumor affects mainly Asians and Native Americans, and is uncommon in Caucasians and Africans, indicating a role of ethnicity in its pathogenesis. The tumor comprises EBV-positive lymphoid cells of diverse morphology and many non-neoplastic inflammatory cells. Necrosis with many apoptotic bodies is a common histological change seen in cases of ENKTL, which can be ascribed to angiocentricity and EBV infection of tumor cells.1
NK and T cells share some phenotypic and functional characteristics. NK cells can express T-cell-associated markers such as CD2, CD3ɛ, CD7, and CD8, but do not express surface CD3 or CD5. Subsets of cytotoxic T cells express the NK-cell-associated markers, CD16, CD56, and CD57. Many activating and inhibitory receptors are expressed in NK cells as well as in subsets of cytotoxic T cells. Such immunophenotypic sharing by NK and T cells blurs the distinction between these cell types.3, 4
Analysis of T-cell receptor gene (TCR) rearrangements is the gold standard to distinguish T from NK cells. The productive rearrangement of the TCR gene in mature T cells distinguishes the two cell types unambiguously.4 TCR gene rearrangement analysis using Southern blot hybridization or nested polymerase chain reaction (PCR) analyses disclosed variable T-cell clonality in ENKTLs ranging from 0 to 27%.5, 6 The BIOMED-2 Concerted Action protocol using multiplex PCR primers has now replaced conventional PCR in assaying gene rearrangements and has higher sensitivity than conventional PCR analyses that use fewer primer sets. Using this technology, monoclonal TCR-γ gene rearrangements were described in 8 of 27 cases of primary cutaneous ENKTLs, which did not express for TCR protein.7, 8
In addition to gene rearrangement analysis, TCR protein detection in tumor cells is useful to distinguish cellular lineages in cases of ENKTL. After completion of productive TCR gene rearrangement, a primary transcript is synthesized that incorporates the constant domain gene, which allows translation of the full-length protein for the TCR chain. Expression of TCR gene transcripts in cases of ENKTL has not yet been studied because of technical difficulties. RNA in situ hybridization (ISH) technology using oligonucleotide probes is now available for formalin-fixed paraffin wax-embedded tissues. Mature T cells express either TCRαβ or TCRγδ proteins on the surface. The αβ T-cell type is identified by the presence of βF1, which marks the constant region of the TCR-β gene. Detection of γδ T cells in formalin-fixed paraffin wax-embedded (FFPE) tissue is now available using recently developed antibodies for the γ- and δ-chains.
The clinical impact of cellular lineages in T-cell lymphomas is controversial. The γδ T-cell phenotype in cutaneous T-cell lymphomas predicts aggressive clinical behavior compared with the αβ T-cell phenotype, and has led to the separation of γδ T-cell lymphomas involving subcutaneous tissue from subcutaneous panniculitis-like T-cell lymphomas, according to the 2008 WHO classification.9 However, in recent studies the clinical impact of cellular lineage by itself has been controversial in cases of cutaneous T-cell lymphomas. The γδ T-cell phenotype is not necessarily associated with bad prognosis, as it can be found in cases with good and bad outcomes.10 On the other hand, in cases of intestinal T-cell lymphoma, the NK-cell lineage predicts a worse prognosis compared with the T-cell lineage, irrespective of the presence of EBV.11 There have been only few reports on the clinical impact of cellular lineage for cases of ENKTL. Ng et al.5 failed to observe any difference in patient survival or clinicopathologic features between true NK-cell tumors and their T-cell counterparts. Takata et al.7 reported that NK-cell cutaneous ENKTLs more frequently showed lymph node involvement—usually CD8-negative and CD56-positive—but no significant difference in overall patient survival was seen between NK- and T-cell lineages.
In this study, we analyzed cellular lineages in cases of ENKTL by gene rearrangement analysis applying BIOMED-2 multiplex assays and immunohistochemistry for TCR types to investigate the clinical impact of cellular lineage. In addition, we evaluated the utility of RNA ISH to detect TCR gene transcripts in determining cellular lineages.
Materials and methods
Cases and Immunohistochemistry
Tissues from 148 cases of ENKTL, nasal type, diagnosed in the Samsung Medical Center from 1996 to 2011, were retrieved. Diagnosis was based on the 2008 WHO classification.1 The histology of each case was reviewed. In all cases, immunohistochemistry was applied for CD3, CD30 (DAKO, Carpinteria, CA, USA), CD4 (Thermo Scientific, Waltham, MA, USA), CD5, CD8, CD56, granzyme B, and for EBV-encoded small RNAs using ISH kits for EBV (Leica Microsystems, Newcastle upon Tyne, UK). TCR gene rearrangement analyses and immunohistochemistry for TCR-βF1 and TCR-cγM1 (Thermo Scientific, Rockford, IL, USA) were performed for virtually all cases. Finally, 108 cases were selected for analysis. In addition, 4 μm sections of normal tonsil and appendix, two cases of enteropathy-associated T-cell lymphoma (EATL) of the TCR-αβ and TCR-γδ phenotypes, and one case of aggressive NK cell leukemia were used to compare the RNAscope assay results with those of immunohistochemistry.
Multiplex PCR Analysis for TCR Gene Rearrangements
TR gene rearrangements were analyzed using a commercial BIOMED-2 multiplex PCR system: an IdentiClone TCR gene γ/β gene clonality assay (InVivoScribe Technologies, San Diego, CA, USA). Tissue sections (5 μm) were deparaffinized with xylene and washed three times with 100% ethanol. Genomic DNA was extracted using ReliaPrep FFPE gDNA extraction kits (Promega, Madison, WI, USA) following the manufacturer’s instructions. Genomic DNA quality was assessed using BIOMED-2 control PCR assay kits (InVivoScribe Technologies), NanoDrop 1000 (Thermo Scientific, Rockford) and Quantus (Promega). Samples with a DNA product size of more than 200 bp were analyzed using BIOMED primers. Those with a DNA product size of less than 200 bp or considered inadequate were excluded.
Genomic DNA was amplified with three multiplex TCR-β gene master mixes (TRB Tubes A, B, C) and two TCR-γ gene master mixes (TRG Tubes A, B). All positive and negative DNA controls, as well as an internal Amplification Control master mix, were run simultaneously. The PCR reaction components and standard program were set up to follow the provider’s protocols. The PCR cycling conditions included initial denaturation at 95 °C for 7 min, 35 cycles with each cycle of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 30 s. For cases with sufficient DNA, each PCR study was carried out in duplicate for 69 and in triplicate for 33 cases, as separate experiments using the same DNA as in the first round or new DNA extracted from the same paraffin block used for the first round. In six cases with insufficient DNA, only single tests were carried out. Gene scanning was carried out on an ABI Prism 3130Xl Sequencer using GeneMapper Software (version 4.0; Applied Biosystems, Foster City, CA, USA).
RNA ISH for TCR Gene Transcripts
Analysis for mRNA expression of the TCR-β or TCR-γ gene was performed using RNAscope assays (Advanced Cell Diagnostics, Hayward, CA, USA) according to the manufacturer’s instructions. FFPE tumor tissues were cut into 4-μm sections and deparaffinized. After pretreatment of the slides, TCR gene probes (TCR-β gene probe targeting the TCR-β constant 1 and 2 genes, catalog No. 433661; TCR-γ gene probe targeting the TCR-γ constant gene, catalog No. 433341) were added to the slides and hybridized in an HybEZ oven (Advanced Cell Diagnostics) for 4 h at 40 °C. Amplification and detection of the signal was performed using RNAscope 2.0 HD detection kits. As a control, RNAscope Control Slide-Human HeLa cell pellets were hybridized with RNAscope control probes.
Determination of Cell Lineages
Cell lineage was determined based on the integrated results of TCR gene rearrangements and immunohistochemistry for TCRs. The algorithm used to determine cell lineage is shown in Figure 1. Cases showing monoclonality by the BIOMED-2 assay were considered as being in the T-cell lineage. Cases with uninterpretable results by the BIOMED-2 assay were divided into T or NK lineages depending on the expression of TCR by immunohistochemistry. Cases that expressed TCR gene transcripts but not the TCR protein were assigned as an undetermined lineage.
Statistical Analysis
The statistical analysis was performed using IBM SPSS (v. 21.0; IBM Corp., Armonk, NY, USA). Correlations between variables were evaluated using Pearson’s r2 test. Univariate and multivariate analysis was performed using Cox proportional hazard regression models (95% confidence interval, CI) with stepwise variable selection. Patient survival was plotted using a Kaplan–Meier curve, and survival curves were compared using the log-rank method. Survival was measured from the date of diagnosis to the last follow-up (which varied); P-values of <0.05 were considered statistically significant.
Results
Clinical Findings
Clinical information on the 108 cases is summarized in Table 1. The primary sites of tumors were the nasal cavity in 67 cases (62%), the extranasal upper aerodigestive tract in 17 (15.7%), and other sites including the skin, soft tissues, and gastrointestinal (GI) tract in 24 (22.3%). Clinical parameters including age, sex, stage at presentation, B-symptoms, and involvement of lymph node and bone marrow were not significantly different among the three groups. However, patients with tumors arising from the nasal cavity showed better performance, normal lactate dehydrogenase (LDH) level, localized stage, and lower International Prognostic Index (IPI) score compared with tumors arising from non-nasal sites including the extranasal upper aerodigestive tract, the skin, soft tissues, and GI tract. Most patients were treated with chemotherapy with or without radiotherapy. Thirty-eight patients received anthracycline-based chemotherapy while another 55 patients were treated with non-anthracycline-based chemotherapy such as dexamethasone, methotrexate, ifosfamide, l-asparaginase, and etoposide (SMILE), ifosfamide, methotrexate, VP-16, and prednisone (IMPS), and etoposide, ifosfamide, cisplatin, and dexamethasone (VIPD). Among the remaining 15 patients, three patients received radiation therapy only and three patients died before treatment. In eight patients, treatment information was not available. The overall median survival was 35.5 months for all patients and 168, 9.5, and 9.5 months for tumors in the nasal region, extranasal upper aerodigestive tract, and other sites, respectively (P=0.000 by log-rank test; Figure 5a).
TCR Gene Rearrangements
Our protocol followed the EuroClonality/BIOMED-2 guidelines for the interpretation and reporting of immunoglobulin/TCR gene clonality12 with some modifications. In short, cases were considered clonal if at least one significant peak (a peak ≥2.5 × the height of the tallest neighboring peak) was detected in one of the TCR-β or -γ gene reactions. The results were divided into two groups: one with a monoclonal peak and the other with equivocal results including cases with no PCR product, a PCR peak that could not be reproduced, or polyclonal peak detected in only some tubes. Among the 108 cases, 44 (40%) showed a reproducible discrete peak and were interpreted as having T-cell monoclonality. Sixty-four cases were uninterpretable; 17 showed polyclonal peaks in some tubes of the TCR-β or TCR-γ gene kits, but no PCR product was observed in other tubes. Twenty-three cases showed no PCR product in any TCR-β gene or -γ gene tubes. Twenty-four cases failed to show any reproducibility of peaks (Supplementary Table S1).
Expression of the TCR Protein
ENKTL expressed TCR-βF1 in 28 cases, TCR-cγM1 in one case, and both forms in one other case (Figure 2). The TCR protein was positive in 30/60 cases of T-ENKTLs (50%) and none of 32 NK-ENKTLs, and 15/40 monoclonal cases (35%) and 15/64 uninterpretable cases (23%) using the BIOMED-2 assay (Supplementary Table S1). ENKTLs usually had non-neoplastic inflammatory cells admixed with neoplastic cells. Because some ENKTLs lacked obvious atypia of tumor cells, we considered both cellular atypia and positive cell count >20% of infiltrating cells to determine the positivity of TCR-βF1. In the TCR-positive sample, the numbers of TCR-positive cells varied, from cases showing TCR-βF1 positivity in only a fraction of tumor cells to cases being positive in the majority of tumor cells (Figure 3). Heterogeneous expression of TCR protein among tumor cells in the same case suggests selective downregulation of TCR expression in a subset of tumor cells.
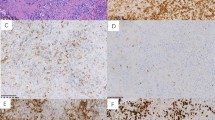
A case of T-lineage ENKTL in testis. (a). The T-cell receptor gene rearrangement assay by using a BIOMED-2 Concerted Action protocol showed a monoclonal peak (a). The tumor comprised of monomorphic medium to large sized tumor cells (b). Virtually all tumor cells were positive for EBV demonstrated by EBER in situ hybridization (c), TCR-βF1 (d), and TCR-CγM1 (e). EBER, EBV-encoded small RNAs; EBV, Epstein–Barr virus; ENKTL, extranodal natural killer/T-cell lymphoma.
T-cell receptor (TCR) expression in T-lineage ENKL. The numbers of TCR-positive neoplastic cells varied, from samples showing positivity in almost all atypical cells (a–c) to samples showing a few positive cells (d–f). EBER in situ hybridization (b,e). TCR-βF1 (c,f). EBER, EBV-encoded small RNAs; ENKTL, extranodal natural killer/T-cell lymphoma.
Expression of Other Markers
CD56 was the most sensitive marker for ENKTL and was expressed in almost all tumor cells. CD56 was expressed in 89 of 96 cases of ENKTL (93%), including 54 of 58 nasal ENKTL (93%), 12 of 15 extranasal upper aerodigestive tract ENKTL (80%), and all 23 ENKTL in other sites (100%). CD56 was positive in 24 of 26 cases of NK-cell ENKTL (92%), 51 of 56 T-cell ENKTL (91%), and all 14 ENKTL of undetermined cell type. CD56 expression did not correlate with clinical parameters or cell lineage. Compared with the number of CD56-positive tumor cells, T-cell markers including CD4, CD5, and CD8 were expressed in only a fraction of the tumor cells and stained positively in many non-neoplastic lymphocytes. Using the cutoff value of 30%, CD4 was expressed in 4 of 56 cases of T-cell ENKTL (7%), CD8 in 11 of 56 cases (20%), and CD5 in 12 of 52 cases (23%). In NK-cell ENKTL, no significant staining of T-cell markers in tumor cells was observed. Using the cutoff value of 10%, CD30 was expressed in 40 of 86 cases (47%), including 24 of 52 nasal ENKTL (46%), 3 of 12 extranasal upper aerodigestive tract ENKTL (25%), and 13 of 22 ENKTL in other sites (59%). Overall 46% of ENKTL cases, including 32, 50, and 56% of NK, T, and undetermined cell types, respectively, expressed CD30 (P>0.05) (Supplementary Table S1).
Expression of TCR Gene Transcripts
Using ISH, the TCR gene transcript was detected in discrete brown dots or conglomerated brown granules that covered the nucleus and cytoplasm. In normal tissue from the tonsil and appendix, TCR-β constant gene mRNA was expressed in most cells in the interfollicular area, and in some cells in the light zone and periphery of the germinal center (Figure 4a). This was consistent with the known distribution of TCR-βF1-positive T cells, indicating staining of αβ T cell (Figure 4b). These cells showed numerous positive signals. An unexpected finding was that one or two discrete, small, positive signals could be observed in the nuclei of most mantle zone cells, and in some centroblasts and centrocytes of the germinal center (Figure 4e). These positive signals in B cells were considered germline transcripts, which have been described in hematopoietic cells.13, 14, 15, 16 TCR-γ constant gene mRNA was expressed in only a few cells scattered in interfollicular, intraepithelial, or intrafollicular areas. These scattered cells each showed numerous signals (Figure 4c). This distribution was compatible with those of TCR-cγM1-positive T cells, and so these cells were interpreted as γδ T cells (Figure 4d). In addition to these γδ T cells, a few cells in germinal centers and primary follicles showed one or two signals in their nuclei, which were interpreted as germline transcripts (Figure 4f). In the two cases of EATL, both showed numerous signals for both TCR-β constant and γ constant gene mRNA, but one was positive only for TCR-βF1, and the other was positive only for TCR-cγM1. One case of aggressive NK cell leukemia showed numerous positive signals for TCR-β constant gene mRNA, but was negative for TCR-γ constant gene mRNA. Taken together, although TCR gene mRNAs were expressed in T cells, this mRNA ISH approach was not useful for distinguishing T cells from other lineages because normal B cells showed germline transcripts and the case of NK-cell leukemia also showed numerous positive signals.
Expression of T-cell receptor gene transcript in normal lymphoid tissue. T-cell receptor-β constant mRNA was expressed in most cells in the interfollicular area, and in some cells in the light zone and periphery of the germinal center. These scattered cells each showed numerous signals (a). This was consistent with the distribution of TCR-βF1-positive T cells (b). T-cell receptor-β constant mRNA signals were observed in the nuclei of most mantle zone cells (e). T-cell receptor-γ constant mRNA was expressed in only a few cells scattered in interfollicular or intrafollicular areas (c). This distribution was compatible with those of TCR-cγM1-positive T cells (d). Only a few TCRγ mRNA-positive cells were observed in normal mantle zone cells (f).
Positive TCR mRNA signals were easily identified in ENKTL cells although they tended to be fewer than in the normal cells from the tonsil or appendix. Among 44 cases of monoclonal ENKTL determined using the BIOMED-2 assay, ISH for TCR-β constant gene mRNA was positive in 13/14 (93%) while that for TCR-γ constant gene was positive in 3/6 cases analyzed using the RNAscope assay. (Supplementary Table S1). On the other hand, among 64 cases with results that could not be interpreted by the BIOMED-2 assay, TCR-β constant gene mRNA was positive in 28/59 analyzed cases (50%) while TCR-γ constant gene mRNA was positive in 6/54 analyzed cases (11%). Five TCR-γ constant gene mRNA-positive cases also expressed TCR-β constant gene mRNA (Supplementary Table S1). TCR-βF1 was expressed in 13/38 cases positive for TCR-β constant gene mRNA (34%) and 3/33 of those negative for TCR-β constant gene mRNA (10%). TCR-cγM1 was expressed in 1/9 cases positive for TCR-γ constant gene mRNA (13%) but in none of 52 cases negative for this mRNA.
Determination of Cell Lineage
The cellular lineage was evaluated in all 108 cases (Figure 1). Forty-four cases with monoclonal TCR gene rearrangements demonstrated by the BIOMED-2 assay were assigned as T-cell type ENKTLs. For those tumors with uninterpretable PCR results, immunohistochemistry for TCR-βF1 and TCR-cγM1 and mRNA ISH were performed. Among these, 16 cases expressed TCR-βF1 or TCR-cγM1 with or without the expression of TCR gene transcripts. These 16 cases were decided to be T-cell type ENKTLs. Sixteen cases were positive for TCR gene transcripts, but were negative for both TCR-βF1 and TCR-cγM1. These cases were defined as belonging to an undetermined lineage. Taken together, T-cell type ENKTLs accounted for 60 cases (56%). Thirty-two cases (30%) were negative for TCR both by RNA ISH and by immunohistochemistry and were considered NK-cell type ENKTLs.
Clinicopathologic Parameters Correlated with Cellular Lineage
As shown in Supplementary Table S2, the cellular lineage of these cases of ENKTL had no significant correlations with various clinical parameters (Supplementary Table S2). NK-cell ENKTL accounted for 39, 12, and 17% of tumors arising from the nasal cavity, extranasal upper aerodigestive tract, and other sites, respectively. The median patient ages were 48, 52, and 43 years for NK-cell ENKTLs, T-cell ENKTLs, and undetermined groups, respectively. The expression of CD56, age, gender, performance status, IPI risk group, B-symptoms, or involvement of bone marrow or lymph nodes showed no differences between NK- and T-cell type ENKTLs. The median patient survival was 38±58 months and 18±16 months for those with NK-cell and T-cell ENKTLs, respectively, with no significant statistical difference between these subgroups (P=0.9) (Figure 5b). Among patients with a localized nasal lymphoma, those with a T-cell ENKTL tended to show a better prognosis (Figure 5c). Six of 20 with an NK-cell type tumor (30%) and 3/26 with a T-cell tumor (12%) died in the first year after diagnosis (P>0.05). The 1-year survival rates for those with T- and NK-cell type tumors were 80 and 63% (P>0.05 by log-rank test), and 71 and 53% at 2 years (P>0.05 by log-rank test). Among 44 patients with localized nasal T-cell-type ENKTLs, 15 expressed TCR protein, and among them 14 expressed TCR-βF1. There was no significant difference in patient survival between cases of localized nasal T-cell ENKTLs that were positive or negative for TCR protein (Figure 5d).
Survival analysis. The overall median survival was 35.5 months for all patients and 168, 9.5, and 9.5 months for tumors in the nasal region, extranasal upper aerodigestive tract, and other sites, respectively (P=0.000 by log-rank test) (a). The median patient survival was 38 and 18 months for those with NK- and T-cell ENKTLs with no significant statistical difference between these subgroups (b). Among patients with a localized nasal lymphoma, those with a T-cell ENKTL tended to show a better prognosis (c). There was no significant difference in patient survival between cases of localized nasal T-cell ENKTLs that were positive or negative for TCR protein (d). ENKTL, extranodal natural killer/T-cell lymphoma; NK, natural killer; TCR, T-cell receptor.
Prognostic Factor Analyses
After a median follow-up of 21 months (range 5 days to 204 months), the median overall survival (OS) was 22 months (95% CI, 0–73.1 months) and the 5-year OS rate was 46%. The clinical factors associated with worse overall survival in the univariate analysis were age >60 years (P=0.024), a non-nasal tumor site (P=0.001), poor performance status (P<0.001), advanced tumor stage >2 (P=0.001), increased LDH level (P=0.001), high-intermediate or high IPI risk group (P<0.001), bone marrow involvement (P=0.001), and the absence of radiotherapy (P<0.001). The chemotherapeutic regimens used showed no significant prognostic influence.
Multivariate analysis showed that a non-nasal tumor site remained as a significant independent poor prognostic factor for OS (hazard ratio (HR), 2.729; 95% CI, 1.263–5.898; P<0.035) (Figure 5a). Other independent prognostic factors for poor OS were age >60 years (HR, 2.110; 95% CI, 1.104–4.035; P=0.024), a high LDH level (HR, 4.732; 95% CI, 2.214–10.112; P=0.001), bone marrow involvement (HR, 4.590; 95% CI, 2.229–9.452; P=0.001), and the absence of radiotherapy (HR, 2.130; 95% CI, 1.140–3.981; P=0.018; Table 2).
Discussion
Here we analyzed the cellular lineages of 108 cases of extranodal ENKTLs by TCR gene rearrangements and immunohistochemistry for TCR proteins. The integrated results demonstrated that these tumors consisted mainly of T-cell types with a minority of NK-cell types. T-lineage ENKTLs mainly expressed TCR-βF1 but not TCR-CγM1. These results are very different from those known for ENKTLs, which have been thought to comprise mainly NK-cell neoplasms and a few cases with γδ T-cell neoplasms.1, 17
Gene rearrangement analysis for T-cell genes is a useful test to identify clonal T-cell proliferation. In cases of ENKTL, previous studies applying conventional PCR techniques showed wide ranges of T-cell clonality ranging from 0 to 71.4%,5, 6, 18, 19 while T-cell clonality ranged from 0 to 26% by BIOMED-2 multiplex PCR analysis.7, 8, 10, 20 The traditional PCR design with a few consensus primers to detect TCR gene rearrangement yields frequent false-negative results because the primers cannot cover extensive sequence variations. On the other hand, a lack of further analysis of PCR products can produce false-positive results. The BIOMED-2 assay, developed by a large collaborative European group, comprises standardized PCR protocols and multiple primer sets to overcome the technical limitations of conventional PCR.21 The sensitivity of this protocol depends on the quality of the target tissue and selection of primer sets. When using fresh-frozen tissue and primer sets for TCR-β and -γ sequences, the BIOMED-2 assay can detect clonality in up to 91% of mature T-cell lymphomas, ranging from 71 to 100% depending on histological subtypes.22 For FFPE samples, the sensitivity has been reported to range from 80 to 98% for TCR.23, 24, 25, 26 ENKTLs are frequently accompanied by necrosis and apoptosis of the tumor cells, leading to degradation of DNA, ineffective PCR reactions, and false-negative results.27 In a study by Pongpruttipan et al.,8 clonal TCR gene rearrangements were documented in only three out of six TCR protein-positive cases. Similarly, our study showed expression of the TCR protein in 15 out of 58 cases with ambiguous PCR results, which comprised 23 with no PCR product, 24 showing peaks that could not be reproduced, and 17 with polyclonality found in only some tubes. These high false-negative results for gene rearrangement analyses in cases of ENKTL suggest that gene rearrangements alone are not sufficient for the analysis of cellular lineages in such tumors. Thus, one should consider integrated data when analyzing the rearrangements and expression levels of TCR genes.
Expression of the TCR genes involves multiple steps. Functional TCR genes are formed by rearrangements of the V and J segments in the α and γ genes, and of the V, D, and J segments in the β and δ genes.13 The α, β, γ, and δ genes also include C segments that do not rearrange. All gene segments between the V(D)J gene segments in the newly formed complex are deleted and a primary transcript is synthesized that incorporates the constant domain gene. To detect TCR transcripts, we performed RNA ISH on slides from FFPE tissues—a newly developed technology. We used oligonucleotide probes directed to a constant region of TCR-β and -γ gene. Because TCR-βF1 and CγM1, which were used in immunohistochemistry, are antibodies produced using a constant region of human TCR-β chain and -γ chain gene as an immunogen, respectively, we expected that TCR gene transcript, in conjunction with immunohistochemistry, mRNA ISH might have provided additional information on the cellular lineage and distribution of αβ T and γδ T cells in tumor tissues. However, the results were disappointing. TCR gene transcripts were expressed in T cells as well as in a case of NK-cell leukemia and in normal B cells. In addition, TCR was expressed in only a portion of TCR gene transcript-positive ENKTL. Thus, TCR mRNA was not a valuable marker in the distinction of cellular lineages. While most transcripts arising from TCR or immunoglobulin gene loci reflect fully rearranged genes, germline transcripts have been identified in cells that have not undergone such gene rearrangements, and these have been thought to be a developmentally regulated lymphoid cell phenomenon.13, 14, 15, 16 In this study, TCR mRNAs expressed in normal B cells and aggressive NK-cell leukemia seemed to be germline transcripts that precluded the utility of using TCR gene transcripts as a marker of T-cell lineage.
The typical immunophenotype of an ENKTL is CD2+, cytoplasmic CD3ɛ+, CD4–, CD5–, CD8–, CD56+, and granzyme B+. Whereas TCR αβ and γδ have usually been reported as negative in ENKTLs,17, 26 recent studies using immunohistochemistry showed variable degrees of TCR expression, ranging from 0 to 23% of these tumors.7, 8, 10, 20 Rodriguez-Pinilla et al.10 showed that ENKTLs primarily affecting the skin or subcutaneous tissue (n=11) were all negative for TCR-γ. Pongpruttipan et al.8 analyzed 67 cases of ENKTL by immunohistochemistry and found that TCR types including βF1, γ, and δ were negative in 70% of cases, positive in only 9%, and indeterminate in 21%. Yamaguchi et al.20 analyzed 32 localized ENKTLs and found that 27 cases (84%) were negative for TCR, and thus considered to be NK-cell tumors; two expressed TCR-βF1 and three expressed TCR-CγM1. On the other hand, Takata et al. analyzed cellular lineages in 51 cases of primary cutaneous ENKTLs. In addition to immunohistochemistry, gene rearrangements were evaluated in those cases with TCR-negative results. Tumors of T-cell origin accounted for 46% of these ENKTLs, with half of them being TCR silent. In the eight cases positive for TCR, the αβ T-cell and γδ T-cell tumors were equal in number.7 In agreement with these data from Takata et al., 50% of the T-lineage ENKTLs lacked TCR in the present study. Absence of TCR expression might be a common phenomenon in T-cell lymphoma and was also reported in EATL.28 Although the mechanism for TCR downregulation remains to be elucidated, lack of TCR in a significant proportion of T-cell type ENKTLs suggests that it might play a role in the pathogenesis of this tumor type. Cytotoxic T(Tc)-cells are key factors in the defense against viral infections. Tc-cells recognize infected cells via the TCR and subsequently kill the target cells by one or more cytotoxic mechanisms. Loss of the TCR protein might enable viral persistence in cases of ENKTL. Investigation of the function of T cells not expressing TCR in ENKTLs might provide an insight to expand our current knowledge on this tumor.
ENKTL is an aggressive disease with a poor prognosis for the patients. The median patient survival is 4.2 years, and 5-year overall survival 46%.29 The prognosis varies depending on the primary site. A significantly better prognosis was reported for patients with an nasal ENKTL, compared with other sites.29, 30, 31 The latter groups had an advanced stage at diagnosis, higher LDH levels, higher IPI scores, worse performance, and a significantly worse response to anthracycline-based chemotherapy.32 A novel regimen containing l-asparaginase (SMILE) had a better effect than anthracycline-based chemotherapy.33, 34 The addition of radiotherapy for early-stage nasal cases yielded survival benefits.35 Biomarkers associated with poor prognosis included the expression of p53 and p63, and high serum levels of interleukin-9, CXCL13, serum VEGF, and survivin.36, 37, 38, 39 EBV-encoded latent membrane protein expression correlated with the phosphorylation of RelA and Akt possibly had a favorable impact on clinical outcome in patients with ENKTLs.20, 40 Among pathology parameters, an adverse prognosis was correlated with a high Ki-67 labeling index and EBV DNA load but not with cellular morphology or the expression of CD56.30, 31, 41 Pongpruttipan et al.8 suggested that patients with a T-cell ENKTL in the upper airway tended to have better survival although this did not reach statistical significance. In our study, the patients with a localized T-cell ENKTL tended to have a better prognosis than those with an NK-cell ENKTL, but the impact of cell lineage on prognosis was not statistically significant. Worthy of note is that the survival of patients with a T-cell ENKTL did not depend on expression of the TCR protein.
ENKTL is an EBV-positive cytotoxic T- or NK-cell lymphoma with a characteristic clinical presentation as defined by the 2008 WHO classification.1 Two recent studies have tried to distinguish EBV-positive cytotoxic T-cell lymphomas from ENKTLs based on the expression of TCR or monoclonal TCR gene rearrangements.42, 43 As shown in this study, the cellular lineage did not have a significant clinical impact for patients with an EBV-positive extranodal cytotoxic cell lymphoma. Thus—at least in clinical contexts—the term ENKTL is suitable to encompass EBV-positive extranodal cytotoxic cell lymphomas irrespective of the presence of T or NK cells in an appropriate clinical setting.
In summary, ENKTLs were mainly derived from αβ T cells expressing TCR-β gene transcripts, but frequently lacked TCR-βF1. Only a third of cases were derived from NK cells and the cell lineage had no significant clinical impact.
References
Chan JKC, Quintanilla-Martinez L, Ferry JA et al. Extranodal NK/T-cell lymphoma, nasal-type. In: Swerdlow SH, Campo E, Harris NL et al (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC: Lyon, 2008, pp 285–288.
Han JH, Ko YH, Kang YK et al. Characteristics of cutaneous lymphomas in Korea according to the new WHO-EORTC classification: report of a nationwide study. Korean J Pathol 2014;48:126–132.
Phillips JH, Hori T, Nagler A et al. Ontogeny of human natural killer (NK) cells: fetal NK cells mediate cytolytic function and express cytoplasmic CD3 epsilon, delta proteins. J Exp Med 1992;175:1055–1066.
Lanier LL . Back to the future—defining NK cells and T cells. Eur J Immunol 2007;37:1424–1426.
Ng SB, Lai KW, Murugaya S et al. Nasal-type extranodal natural killer/T-cell lymphomas: a clinicopathologic and genotypic study of 42 cases in Singapore. Mod Pathol 2004;17:1097–1107.
Ohsawa M, Nakatsuka S, Kanno H et al. Immunophenotypic and genotypic characterization of nasal lymphoma with polymorphic reticulosis morphology. Int J Cancer 1999;81:865–870.
Takata K, Hong ME, Sitthinamsuwan P et al. Primary cutaneous NK/T-cell lymphoma, nasal type and CD56-positive peripheral T-cell lymphoma: a cellular lineage and clinicopathologic study of 60 patients from Asia. Am J Surg Pathol 2015;39:1–12.
Pongpruttipan T, Sukpanichnant S, Assanasen T et al. Extranodal NK/T-cell lymphoma, nasal type, includes cases of natural killer cell and αβ γδ, and αβ/γδ T-cell origin: a comprehensive clinicopathologic and phenotypic study. Am J Surg Pathol 2012;36:481–499.
Jaffe ES, Gaulard P, Ralfkiaer E et al. Subcutaneous panniculitis-like T-cell lymphoma. In: Swerdlow SH, Campo E, Harris NL et al (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC: Lyon, 2008, pp 294–295.
Rodriguez-Pinilla SM, Ortiz-Romero PL, Monsalvez V et al. TCR-gamma expression in primary cutaneous T-cell lymphomas. Am J Surg Pathol 2013;37:375–384.
Chuang SS, Chang ST, Chuang WY et al. NK-cell lineage predicts poor survival in primary intestinal NK-cell and T-cell lymphomas. Am J Surg Pathol 2009;33:1230–1240.
Langerak AW, Groenen PJ, Bruggemann M et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia 2012;26:2159–2171.
Soloff RS, Wang TG, Lybarger L et al. Transcription of the TCR-beta locus initiates in adult murine bone marrow. J Immunol 1995;154:3888–3901.
Abbey JL, Karsunky H, Serwold T et al. Expression of TCR-Vβ peptides by murine bone marrow cells does not identify T-cell progenitors. J Cell Mol Med 2015;19:1956–1964.
Abbey JL, O’Neill HC . Expression of T-cell receptor genes during early T-cell development. Immunol Cell Biol 2008;86:166–174.
Wang TG, Lybarger L, Soloff R et al. Pre-thymic transcription of TCR genes by adult murine bone marrow cells. Mol Immunol 1996;33:957–964.
Cheuk W, Chan JKC. NK-cell neoplasms. In: Jaffe ES, Harris NL, Vardiman JW et al. (eds). Hematopathology. Elsevier: St. Louis, Missouri, 2011, pp 473–491..
Ko YH, Ree HJ, Kim WS et al. Clinicopathologic and genotypic study of extranodal nasal-type natural killer/T-cell lymphoma and natural killer precursor lymphoma among Koreans. Cancer 2000;89:2106–2116.
Pongpruttipan T, Pongtongcharoen P, Sukpanichnant S . Mature T-cell and NK-cell lymphomas in Thailand: an analysis of 71 cases. J Med Assoc Thai 2011;94:743–748.
Yamaguchi M, Takata K, Yoshino T et al. Prognostic biomarkers in patients with localized natural killer/T-cell lymphoma treated with concurrent chemoradiotherapy. Cancer Sci 2014;105:1435–1441.
van Dongen JJ, Langerak AW, Bruggemann M et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003;17:2257–2317.
Bruggemann M, White H, Gaulard P et al. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies. Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia 2007;21:215–221.
Chuang SS, Lin CN, Shen FC et al. Detecting clonal rearrangement in non-Hodgkin’s lymphomas in Taiwan by polymerase chain reaction. Leuk Lymphoma 2003;44:117–121.
Chen YL, Su IJ, Cheng HY et al. BIOMED-2 protocols to detect clonal immunoglobulin and T-cell receptor gene rearrangements in B- and T-cell lymphomas in southern Taiwan. Leuk Lymphoma 2010;51:650–655.
Kim Y, Choi YD, Choi C et al. Diagnostic utility of a clonality test for lymphoproliferative diseases in Koreans using the BIOMED-2 PCR assay. Korean J Pathol 2013;47:458–465.
Liu H, Bench AJ, Bacon CM et al. A practical strategy for the routine use of BIOMED-2 PCR assays for detection of B- and T-cell clonality in diagnostic haematopathology. Br J Haematol 2007;138:31–43.
Emile JF, Boulland ML, Haioun C et al. CD5-CD56+ T-cell receptor silent peripheral T-cell lymphomas are natural killer cell lymphomas. Blood 1996;87:1466–1473.
Tan SY, Chuang SS, Tang T et al. Type II EATL (epitheliotropic intestinal T-cell lymphoma): a neoplasm of intra-epithelial T-cells with predominant CD8αα phenotype. Leukemia 2013;27:1688–1696.
Li S, Feng X, Li T et al. Extranodal NK/T-cell lymphoma, nasal type: a report of 73 cases at MD Anderson Cancer Center. Am J Surg Pathol 2013;37:14–23.
Lee J, Park YH, Kim WS et al. Extranodal nasal type NK/T-cell lymphoma: elucidating clinical prognostic factors for risk-based stratification of therapy. Eur J Cancer 2005;41:1402–1408.
Barrionuevo C, Zaharia M, Martinez MT et al. Extranodal NK/T-cell lymphoma, nasal type: study of clinicopathologic and prognosis factors in a series of 78 cases from Peru. Appl Immunohistochem Mol Morphol 2007;15:38–44.
Liu QF, Wang WH, Wang SL et al. Immunophenotypic and clinical differences between the nasal and extranasal subtypes of upper aerodigestive tract natural killer/T-cell lymphoma. Int J Radiat Oncol Biol Phys 2014;88:806–813.
Kim SJ, Yang DH, Kim JS et al. Concurrent chemoradiotherapy followed by L-asparaginase-containing chemotherapy, VIDL, for localized nasal extranodal NK/T cell lymphoma: CISL08-01 phase II study. Ann Hematol 2014;93:1895–1901.
Kwong YL, Kim WS, Lim ST et al. SMILE for natural killer/T-cell lymphoma: analysis of safety and efficacy from the Asia Lymphoma Study Group. Blood 2012;120:2973–2980.
Au WY, Weisenburger DD, Intragumtornchai T et al. Clinical differences between nasal and extranasal natural killer/T-cell lymphoma: a study of 136 cases from the International Peripheral T-Cell Lymphoma Project. Blood 2009;113:3931–3937.
Ye Z, Cao Q, Niu G et al. p63 and p53 expression in extranodal NK/T cell lymphoma, nasal type. J Clin Pathol 2013;66:676–680.
Zhang J, Wang WD, Geng QR et al. Serum levels of interleukin-9 correlate with negative prognostic factors in extranodal NK/T-cell lymphoma. PLoS One 2014;9:e94637.
Kim SJ, Ryu KJ, Hong M et al. The serum CXCL13 level is associated with the Glasgow Prognostic Score in extranodal NK/T-cell lymphoma patients. J Hematol Oncol 2015;8:49.
Kim SJ, Hong M, Do IG et al. Serum survivin and vascular endothelial growth factor in extranodal NK/T-cell lymphoma, nasal type: implications for a potential new prognostic indicator. Haematologica 2015;100:e106–e109.
Kanemitsu N, Isobe Y, Masuda A et al. Expression of Epstein-Barr virus-encoded proteins in extranodal NK/T-cell lymphoma, nasal type (ENKL): differences in biologic and clinical behaviors of LMP1-positive and -negative ENKL. Clin Cancer Res 2012;18:2164–2172.
Li YX, Wang H, Feng XL et al. Immunophenotypic characteristics and clinical relevance of CD56+ and CD56- extranodal nasal-type natural killer/T-cell lymphoma. Leuk Lymphoma 2011;52:417–424.
Yu WW, Hsieh PP, Chuang SS . Cutaneous EBV-positive γδ T-cell lymphoma vs extranodal NK/T-cell lymphoma: a case report and literature review. J Cutan Pathol 2013;40:310–316.
Pan ST, Chang WS, Murphy M et al. Cutaneous peripheral T-cell lymphoma of cytotoxic phenotype mimicking extranodal NK/T-cell lymphoma. Am J Dermatopathol 2011;33:e17–e20.
Acknowledgements
This study was supported by a grant from the National Strategic Coordinating Center for Clinical Research, the Korean Ministry of Health & Welfare (A102065), and by the National Research Foundation of Korea (NRF-2014R1A2A2A 01007826).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Hong, M., Lee, T., Young Kang, S. et al. Nasal-type NK/T-cell lymphomas are more frequently T rather than NK lineage based on T-cell receptor gene, RNA, and protein studies: lineage does not predict clinical behavior. Mod Pathol 29, 430–443 (2016). https://doi.org/10.1038/modpathol.2016.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.47
This article is cited by
-
Updates in the Classification of T-cell Lymphomas and Lymphoproliferative Disorders
Current Hematologic Malignancy Reports (2023)
-
RETRACTED ARTICLE: Extranodal NK/T-Cell Lymphoma, Nasal Type in Guatemala: An 86-Case Series Emphasizing Clinical Presentation and Microscopic Characteristics
Head and Neck Pathology (2019)