Abstract
Giant cell tumors of bone are locally aggressive bone neoplasms with a predilection for young adults. Histologically, they are composed of histiocytoid to spindled mononuclear cells, admixed with numerous large osteoclastic giant cells. Giant cell tumors of soft tissue are rare tumors that bear striking histological resemblance to giant cell tumors of bone and might be regarded as a soft tissue analog thereof. Point mutations of the H3F3A gene (coding for a histone H3.3 protein) at the Gly34 codon, mostly G34W resulting from a GGG>TGG nucleotide change, have recently been identified in a vast majority of giant cell tumors of bone. To delineate the possible pathogenic linkage between both tumor types, we analyzed the H3F3A genotypes in a series of 15 giant cell tumors of soft tissue by Sanger sequencing and found no mutation in any case. We then sequenced cognate histone H3 genes with an identical nucleotide sequence (‘GGG’) at the codon Gly34, including the H3F3B, H3F3C, HIST2H3A, HIST2H3C, and HIST2H3D genes, and no somatic mutation was detected. These results reveal that giant cell tumors of soft tissue are probably genetically distinct from their bone counterparts and suggest that they might be pathogenically unrelated. Given the prominence of non-neoplastic cells in these tumors and the limitations of the current study, however, analyses using more sensitive techniques might be required to solve the issue.
Similar content being viewed by others
Main
Giant cell tumor of bone is a locally aggressive neoplasm that represents 5% of all primary bone tumors and has a predilection for long bones and vertebrae of young adults.1 Histologically, giant cell tumor of bone is composed of macrophage-like to spindled mononuclear cells, usually interspersed with numerous large osteoclasts. A cytogenetic hallmark of giant cell tumor of bone is the presence of telomeric associations.2 Importantly, it has been shown that >80% of giant cell tumors harbored H3F3A mutations at the codon Gly34, about 95% of which were p.G34W resulting from a GGG>TGG nucleotide alteration.3, 4, 5, 6 Given their high prevalence in giant cell tumor of bone and near non-existence among potential mimics, H3F3A G34 mutations could serve as a useful diagnostic adjunct.3, 4, 7
Giant cell tumor of soft tissue is histologically similar to giant cell tumor of bone and, as the name indicates, might be regarded as the soft tissue analog of the latter.8, 9, 10, 11 Clinically, similar to its osseous counterpart, giant cell tumor of soft tissue is considered a tumor of low malignant potential, with a tendency to local recurrence while rarely metastasizing. Furthermore, telomeric associations have also been characterized in a single case.12 To determine whether giant cell tumor of soft tissue is similar to its osseous counterpart at the genetic level as well, we analyzed a group of cases for the genotypes of H3F3A and the cognate histone H3 genes containing identical codon 34 sequences (ie, ‘GGG’).
Materials and methods
Tumor Samples
Pathology materials of giant cell tumors of soft tissue, diagnosed between 2010 and 2015, were collected from the consultation files of one of the authors (CDMF). The histology was reviewed and confirmed by all the authors.
DNA Extraction
Tumor tissue, and adjacent normal tissue from selective cases, was dissected from the paraffin tissue slides. DNA was then extracted from the dissected tissue using the BiOstic FFPE Tissue RNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA) following the manufacturer’s instructions.
PCR and Sanger Sequencing
The extracted DNA was subjected to PCR and bidirectional sequencing, as previously described,13 using specific primers targeting the N-terminal parts of H3F3A, H3F3B, and H3F3C, as well as the whole coding regions of HIST2H3A/C (both genes have identical UTRs and coding sequences and shared the same primers) and HIST2H3D, respectively. Table 1 demonstrates the primer sequences. The electropherograms of H3F3A G34W and H3F3B K36M mutations from a giant cell tumor of bone and a chondroblastoma, respectively, using the same primer sets served as positive controls (Supplementary Figure S1).
Results
Clinicopathological Features of the Study Group
A total of 15 giant cell tumors of soft tissue were collected. The patients’ ages ranged from 5 to 89 years (median 63 years). Eleven were female and four were male. The tumor locations included the distal upper extremity (six patients), distal lower extremity (four), thigh (two), arm (one), trunk (one), and face (one). Most tumors were centered on the subcutis. Follow-up data were not collected.
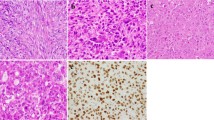
Histologically, the tumors were well circumscribed and showed a striking growth pattern of multiple nodules in 12 cases, separated by sclerotic and often hemosiderotic septa (Figure 1a). The cellular nodules were composed of mononuclear histiocytoid to plump spindle cells and osteoclastic giant cells with rich vasculature (Figure 1b). Both the mononuclear cells and giant cells had similarly round to ovoid nuclei with vesicular chromatin and small distinct nucleoli (Figure 1c). Osseous metaplasia was present in 7 cases (Figures 1a). Mitotic figures ranged from 2 to 18 (median: 5) per 10 high-power fields. Table 2 demonstrates the clinical and pathological information.
The histological features of giant cell tumor of soft tissue. (a) The tumor shows a striking multinodular growth pattern, with the tumor nodules separated by sclerotic bands with hemosiderin deposition. Osseous metaplasia is quite often observed (right lower field). (b and c) The major cell types are histiocytoid mononuclear cells admixed with osteoclastic giant cells; both cell types have morphologically similar nuclei that show vesicular chromatin and distinct nucleoli. Rich vasculature is also noted.
Genotypes of Histone H3 Genes
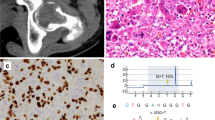
All cases were shown by direct sequencing to harbor wild-type H3F3A and H3F3B genes. Specifically, no mutation was observed at codon Lys27, Gly34, or Lys36 of either gene (Figures 2a and b).
Sufficient DNA remained for subsequent analyses in 14 cases. Of these, 11 cases had a wild-type H3F3C gene. The other three cases harbored a p.H38P single-nucleotide polymorphism (rs3759295), which was also observed in the respective normal DNA (Figure 2c). Of the 11 cases where the PCR was successful, the HIST2H3A/C and HIST2H3D genes were wild type in all, except for 1 case that had a homozygous p.S87S synonymous variant in the HIST2H3A/C genes (Figure 2d). Specifically, no mutation was found at codon Gly34 in any of these cognate histone H3 genes. See Table 2 for details of the sequencing results.
Discussion
Histones are nuclear proteins that function to package the genome into chromatin units known as nucleosomes. Modifications of histones are central to chromatin remodeling and gene expression regulation, thus being critical to a number of cellular processes.14, 15 Dysregulation of histone modifications has prominent implications in cancer formation.15, 16 Recently, recurrent mutations involving the codons Lys27, Lys36, and Gly34 of the histone variant H3.3 (encoded by the H3F3A and H3F3B genes) have been identified in pediatric glial tumors, chondroblastoma, and giant cell tumor of bone, respectively.3, 17, 18 These mutations probably alter the global landscape of epigenetic modifications through interfering with histone methyltransferases, such as EZH2.19, 20, 21
Giant cell tumors of bone and of soft tissue are similar in multiple aspects. The histological resemblance probably accounts for the shared terminology, as both tumors are characterized by striking number of large osteoclastic giant cells and histiocytoid mononuclear cells. The mononuclear cells in both tumors have been shown to exhibit an osteoblastic phenotype.22 Both often have rich vasculature and can show prominent osseous metaplasia. Clinically, both tumors are considered to be locally aggressive. Cytogenetically, giant cell tumor of bone typically showed clonal and non-clonal telomeric associations, which has also been reported in one giant cell tumor of soft tissue.2, 12 On the other hand, however, there is no ignoring some differences between these two entities. The oftentimes prominent multinodular pattern of soft tissue giant cell tumor is not a feature of the bone counterpart. In contrast to giant cell tumor of bone, which mostly afflicts young to middle-aged adults, giant cell tumor of soft tissue tends to occur in patients with a wider range of age.9, 10, 11, 23 It is also noteworthy that the phenomenon of telomeric associations is not specific and can be found in a variety of neoplasms.24, 25, 26, 27, 28 Therefore, the identification of telomeric associations in just one case of soft tissue giant cell tumor can hardly be regarded as solid evidence that both tumors belong to one unified entity.
In view of the high prevalence of H3F3A Gly34 mutations in giant cell tumor of bone, we carried out genotyping of the H3F3A gene on the soft tissue counterpart to delineate the genetic linkage between these two tumors. The current data revealed no mutation of the H3F3A gene in any of the 15 giant cell tumors of soft tissue, demonstrating a disparity between these tumor types at the genetic level. However, it did not exclude the possible involvement of alternative histone proteins in the pathogenesis of giant cell tumor of soft tissue. As G34W accounts for a vast majority of the H3F3A mutations in giant cell tumor of bone, it is reasonable to postulate that a mutant tryptophan at codon 34 might have virtually irreplaceable biological significance that is specific to this tumor type. Among the codons Gly34 of all histone H3 proteins (coded by GGG, GGC, or GGT), only those with the GGG sequence can be conveniently converted to tryptophan (coded by TGG) with a single nucleotide substitution. This constituted the rationale to scrutinize all histone H3 genes with a codon 34 composed of GGG sequence other than H3F3A, ie, H3F3B, H3F3C, HIST2H3A/C, and HIST2H3D. No somatic mutation was disclosed in these genes. These results further argue for the separation of giant cell tumors of bone and of soft tissue into two distinct entities.
The current study has limitations. First, despite the meticulous dissection to enrich the tumor tissue, the presence of numerous admixed non-neoplastic cells (such as the osteoclastic giant cells) was inevitable and could decrease the sensitivity of mutation detection by Sanger sequencing. Second, as the HIST2H3A and HIST2H3C genes have identical coding and flanking sequences and therefore share the same primer set, the sensitivity of detecting mutations in either gene is further lowered. Finally, the possibility remains that giant cell tumor of soft tissue might harbor aberrations of other histone proteins or associated factors involved in the epigenetic pathways, thus imparting a link to its osseous counterpart at the molecular level. More comprehensive or sensitive techniques, such as genome-wide or deep next-generation sequencing, may be required to dissect these issues.
In conclusion, we have analyzed the genotypes of H3F3A and other cognate histone H3 genes in a series of 15 giant cell tumors of soft tissue. The results suggest that giant cell tumor of soft tissue might be genetically different from its osseous counterpart, and these two tumor types might therefore be better regarded as two distinct entities. More studies are warranted to shed light on the pathogenesis of giant cell tumor of soft tissue.
References
Athanasou NA, Bansal M, Forsyth R et al, Giant cell tumour of bone. In: Fletcher CDM, Bridge JA, Hogendoorn PCW et al, (eds).World Health Organization Classification of Tumours of Soft Tissue and Bone. IARC Press: Lyon, France, 2013, pp321–324.
Bridge JA, Neff JR, Mouron BJ . Giant cell tumor of bone. Chromosomal analysis of 48 specimens and review of the literature. Cancer Genet Cytogenet 1992; 58: 2–13.
Behjati S, Tarpey PS, Presneau N et al, Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 2013; 45: 1479–1482.
Cleven AH, Hocker S, Briaire-de Bruijn I et al, Mutation analysis of H3F3A and H3F3B as a diagnostic tool for giant cell tumor of bone and chondroblastoma. Am J Surg Pathol 2015; 39: 1576–1583.
Girolami I, Mancini I, Simoni A et al, Denosumab treated giant cell tumour of bone: a morphological, immunohistochemical and molecular analysis of a series. J Clin Pathol 2016; 69: 240–247.
Al-Ibraheemi A, Inwards CY, Zreik RT et al, Histologic spectrum of giant cell tumor (GCT) of bone in patients 18 years of age and below: a study of 63 patients. Am J Surg Pathol 2016; 40: 1702–1712.
Gomes CC, Diniz MG, Amaral FR et al, The highly prevalent H3F3A mutation in giant cell tumours of bone is not shared by sporadic central giant cell lesion of the jaws. Oral Surg Oral Med Oral Pathol Oral Radiol 2014; 118: 583–585.
Oliveira AM. Giant cell tumour of soft tissue. In: Fletcher CDM, Bridge JA, Hogendoorn PCW et al, (eds).World Health Organization Classification of Tumours of Soft Tissue and Bone. IARC Press: Lyon, France, 2013, pp 106–107.
Folpe AL, Morris RJ, Weiss SW . Soft tissue giant cell tumor of low malignant potential: a proposal for the reclassification of malignant giant cell tumor of soft parts. Mod Pathol 1999; 12: 894–902.
Oliveira AM, Dei Tos AP, Fletcher CD et al, Primary giant cell tumor of soft tissues: a study of 22 cases. Am J Surg Pathol 2000; 24: 248–256.
O'Connell JX, Wehrli BM, Nielsen GP et al, Giant cell tumors of soft tissue: a clinicopathologic study of 18 benign and malignant tumors. Am J Surg Pathol 2000; 24: 386–395.
Guo H, Garcia RA, Perle MA et al, Giant cell tumor of soft tissue with pulmonary metastases: pathologic and cytogenetic study. Pediatr Dev Pathol 2005; 8: 718–724.
Lee JC, Su SY, Changou CA et al, Characterization of FN1-FGFR1 and novel FN1-FGF1 fusion genes in a large series of phosphaturic mesenchymal tumors. Mod Pathol 2016; 29: 1335–1346.
Kouzarides T . Chromatin modifications and their function. Cell 2007; 128: 693–705.
Jones PA, Baylin SB . The epigenomics of cancer. Cell 2007; 128: 683–692.
Liu F, Wang L, Perna F et al, Beyond transcription factors: how oncogenic signalling reshapes the epigenetic landscape. Nat Rev Cancer 2016; 16: 359–372.
Schwartzentruber J, Korshunov A, Liu XY et al, Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012; 482: 226–231.
Wu G, Broniscer A, McEachron TA et al, Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012; 44: 251–253.
Chan KM, Fang D, Gan H et al, The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 2013; 27: 985–990.
Lewis PW, Muller MM, Koletsky MS et al, Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013; 340: 857–861.
Bender S, Tang Y, Lindroth AM et al, Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 2013; 24: 660–672.
Lau YS, Sabokbar A, Gibbons CL et al, Phenotypic and molecular studies of giant-cell tumors of bone and soft tissue. Hum Pathol 2005; 36: 945–954.
Salm R, Sissons HA . Giant-cell tumours of soft tissues. J Pathol 1972; 107: 27–39.
Fitzgerald PH, Morris CM . Telomeric association of chromosomes in B-cell lymphoid leukemia. Hum Genet 1984; 67: 385–390.
Mandahl N, Heim S, Kristoffersson U et al, Telomeric association in a malignant fibrous histiocytoma. Hum Genet 1985; 71: 321–324.
Kovacs G, Muller-Brechlin R, Szucs S . Telomeric association in two human renal tumors. Cancer Genet Cytogenet 1987; 28: 363–366.
Sawyer JR, Roloson GJ, Bell JM et al, Telomeric associations in the progression of chromosome aberrations in pediatric solid tumors. Cancer Genet Cytogenet 1996; 90: 1–13.
Paz-y-Mino C, Sanchez ME, Del Pozo M et al, Telomeric association in women with breast and uterine cervix cancer. Cancer Genet Cytogenet 1997; 98: 115–118.
Acknowledgements
This work was supported by the research fund from National Taiwan University Hospital, Taipei, Taiwan (NTUH 105-S3041) to Dr Jen-Chieh Lee. We thank Mr. Yung-Chuan Chung for the technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, JC., Liang, CW. & Fletcher, C. Giant cell tumor of soft tissue is genetically distinct from its bone counterpart. Mod Pathol 30, 728–733 (2017). https://doi.org/10.1038/modpathol.2016.236
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.236
This article is cited by
-
Establishment and characterization of the NCC-GCTB4-C1 cell line: a novel patient-derived cell line from giant cell tumor of bone
Human Cell (2022)
-
Recurrent novel HMGA2-NCOR2 fusions characterize a subset of keratin-positive giant cell-rich soft tissue tumors
Modern Pathology (2021)
-
Edgetic perturbation signatures represent known and novel cancer biomarkers
Scientific Reports (2020)
-
What is new about the molecular genetics in matrix-producing soft tissue tumors? -The contributions to pathogenetic understanding and diagnostic classification
Virchows Archiv (2020)
-
Evaluation of giant cell tumors by diffusion weighted imaging–fractional ADC analysis
Skeletal Radiology (2019)