Abstract
The biological role of quiescin sulfhydryl oxidase 1 (QSOX1) in tumor development is not well known, and its relation to breast cancer progression and prognosis is controversial. Here, our aim was to study the expression pattern and prognostic impact of QSOX1 in breast cancer, in relation to molecular subgroups and tumor cell proliferation. We examined a population-based series as part of the prospective Norwegian Breast Cancer Screening Program, including all women (50–69 years) diagnosed with breast cancer in one county of Norway during 1996–2003. QSOX1 expression was assessed by immunohistochemistry on tissue microarrays (n=458). Median follow-up time was 13 years. High expression of QSOX1 protein was associated with features of poor prognosis including high histologic grade, hormone receptor negativity, HER2 positivity, and increased tumor cell proliferation. High QSOX1 expression was further associated with reduced breast cancer-specific survival in both univariate and multivariate analysis, independent of molecular subtypes. High QSOX1 expression is a strong and independent factor of reduced survival in breast cancer, also reflected by elevated levels in more aggressive molecular subgroups. QSOX1 expression may represent a biomarker for aggressive disease and a potential treatment target.
Similar content being viewed by others
Main
Breast cancer is the most common type of malignant tumors among women, and despite advances in diagnosis and therapy, it is a major cause of illness and death. Thus, it is important to explore the clinical relevance of novel biomarkers and possible treatment targets.
Quiescin sulfhydryl oxidase (QSOX1) is an enzyme mainly localized to the Golgi apparatus.1, 2 It catalyzes disulfide bond formation in proteins during folding, with concurrent reduction of oxygen to hydrogen peroxide.3 QSOX1 is also secreted, and highest expression has been shown in secretory cells.4
Although previously associated with fibroblast quiescence,5 it has become increasingly clear that QSOX1 is also related to malignancy.6 Elevated QSOX1 expression has been shown in prostate, breast, and pancreatic cancer,7, 8, 9, 10 and high levels have been associated with an invasive phenotype in breast and pancreatic cell lines.11, 12 Secreted QSOX1 may also have an important role in creating a pro-invasive environment.2
However, the prognostic impact of QSOX1 expression in breast cancer remains controversial. One report showed that high QSOX1 mRNA expression was associated with an adverse prognosis in Luminal B breast cancer.11 In contrast to this, high QSOX1 mRNA expression has been correlated with favorable outcome.13 Thus, the value of QSOX1 expression as a prognostic biomarker in breast cancer remains undetermined.
On this background, our aim was to study the expression of QSOX1 in a well-defined population-based series of breast cancers, with clinico-pathologic associations and prognostic impact as end points. Second, we wanted to assess QSOX1 expression in relation to tumor cell proliferation and molecular subgroups of breast cancer.
Materials and methods
Patient Series
A population-based series of 534 breast carcinomas was studied.14 In brief, women aged 50–69 years diagnosed with primary breast carcinoma during 1996–2003 as part of the prospective Norwegian Breast Cancer Screening Program were included. The patients received treatment according to standard protocols at the time. Follow-up information was provided by the Norwegian Cause of Death Registry, and can be considered accurate and complete. Last date of follow-up was 31 December 2011. Outcome data include survival status, survival time, and cause of death. During the follow-up period, 79 patients (15%) died from breast carcinoma, and 62 (12%) died from other causes. The median follow-up was 13 years calculated by the reverse Kaplan–Meier method. This study was approved by the Western Regional Committee for Medical and Health Research Ethics, REC West (REK 2012/1704).
Clinico-Pathological Variables
The following variables were recorded: age at diagnosis, tumor diameter, histologic type, histologic grade, lymph node status, and hormonal receptor status. Mitotic count was done separately and reported as mitoses per mm2. Furthermore, selected results of previously investigated biomarkers (HER2, Ki67) were included.14 Of 534 patients in the complete cohort, tissue microarrays were available for 458 patients (86%).
Tissue Microarrays
H&E-stained slides were used for tumor verification, as previously described.14 In brief, 1.0 mm cores in triplicate were punched and mounted into a recipient paraffin block using a semiautomated precision instrument (Minicore 3, Tissue Arrayer, Alphelys, France). Areas of high tumor purity, the tumor periphery and the highest histologic grade were selected. 190 cases had previously been processed;15 from these cases three tissue cores with a diameter of 0.6 mm were obtained by a different instrument (Beecher Instruments, Silver Spring, MD, USA).
Immunohistochemistry
Staining was performed on thin tissue microarray sections (5 μm) from formalin-fixed and paraffin-embedded tumor tissue. The sections were de-waxed with xylene/ethanol before target retrieval in a microwave oven (6th Sense) in Target Retrieval Solution (pH=6) for 20 min. Endogenous peroxidase activity was blocked. Staining procedures were performed using a DAKO autostainer.
QSOX1
Sections were incubated for 60 min at room temperature (22 °C) with a rabbit polyclonal QSOX1 antibody diluted 1:100 (Proteintech Group, Inc., Rosemont, IL, USA), corresponding to previously published protocols aside from a small adjustment of concentration.10, 11, 12, 13 Detection was done by Dako EnVision rabbit for 30 min.
CK5/6
Staining procedures were performed using the K4061/Envision Dual Link System (rabbit+mouse). Slides were incubated for 30 min with a mouse monoclonal antibody (Dako M7237, clone D5/16 B4) at a 1:200 dilution.16
P-cadherin
P-cadherin staining was obtained by incubating sections for 60 min with a mouse monoclonal antibody (clone 56) from BD Biosciences (San Jose, CA, USA) at a dilution of 1:400.15, 17 Detection was done by Dako Envision mouse for 30 min. Sections were developed in DAB chromogen (Dako REAL DAB+chromogen) and counterstained with Dako REAL hematoxylin.
For QSOX1, sections from a breast carcinoma known to yield positive staining were used as controls. For CK5/6 and P-cadherin; tonsillar tissue was used as positive control. Negative controls were obtained by replacing the primary antibody with Tris-buffered saline.
Evaluation of Staining
Tissue microarrays were examined and scored by one pathologist (GK) blinded to patient characteristics and outcome. Staining was recorded by use of a semi-quantitative and subjective grading system as previously described.18, 19 In brief, a staining index (values 0–9) was calculated as a product of staining intensity (0–3) and proportion of tumor cells showing a positive reaction (0=no staining, 1<10%, 2=10–50% and 3>50%). In subsequent statistical analyses, cutoff points were mainly based upon median or quartile values, also considering the frequency distribution for each marker. Categories with similar survival estimates were merged.
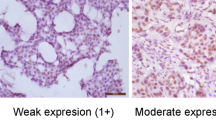
QSOX1 staining was cytoplasmic with a granular, dot-like appearance. In some sections a weak, cytoplasmic staining of normal breast epithelium, endothelium, stromal cells, and lymphocytes was noted. There are no validated QSOX1 cutoff points based on immunohistochemical staining. In this study, QSOX1 low expression encompassed carcinomas showing weak to moderate staining and/or more focal staining (staining index 0–6). QSOX1 high expression was denoted in cases demonstrating strong cytoplasmic staining intensity in 50% or more of the tumor cells (staining index 9).
CK5/6 positivity was cytoplasmic. P-cadherin staining was mainly membranous, although some cytoplasmic staining was observed in cases showing strong overall staining.
For CK5/6, a positive staining was defined as staining index>0, whereas for P-cadherin, staining index>3 was used, as previously described.15, 20
One observer (GK) assessed the series twice, with good intra-observer reproducibility, Kappa-value 0.89, P<0.001. For QSOX1, a subset (n=66) was scored by two observers (GK, IS) with substantial concordance (kappa-value 0.83, P<0.001). For P-cadherin and CK5/6, good inter-observer agreement has previously been shown.15
Surrogate Definitions of Molecular Subclasses of Breast Cancer
Molecular classes were defined as proposed by St Gallen 2013,21 with minor modifications. Luminal A-like: ER and/or PR positive, HER2 negative, Ki67<20%; Luminal B-like (HER2 negative): ER positive, HER2 negative, Ki67≥20% and/or PR negative; Luminal B-like (HER2 positive): ER positive and HER2 overexpressed or amplified, any Ki67, any PR; HER2 positive (non-luminal): HER2 overexpressed or amplified, ER and PR absent; Triple negative: ER and PR absent, HER2 negative. Our modifications include the use of an ER and PR cutoff point of 10% according to national guidelines at that time. A small subgroup (11 cases) in our series showed ER negativity, PR positivity and HER2 negativity; these were not possible to classify by the 2013 criteria. Based on the St Gallen 201122 we decided to categorize these in the luminal subgroup.
Statistical Methods
Analyses were performed using IBM SPSS Statistics for Windows, Version 21.0. (Armonk, NY, USA: IBM Corp). Statistical significance was assessed at the two-sided 5% level. Associations between categorical variables were evaluated by Pearson’s χ2-test. The Cohen’s kappa measure was used to assess the agreement of categorical scores. Ordinal variables were compared between two or more groups using Mann–Whitney U or Kruskal–Wallis H-tests.
For survival analyses, the end point was breast cancer-specific survival, defined as the time in months from diagnosis until death from breast cancer. Patients with missing data were excluded from analyses. Univariate survival analyses were performed using the product-limit procedure (Kaplan–Meier method), and differences between categories were estimated by the log-rank test. Patients who died from other causes were censored. Univariate and multivariate survival analyses were conducted using Cox’ proportional hazards methods (likelihood ratio test). Multivariate analyses were adjusted for standard prognostic factors including tumor size, histologic grade, nodal status and age. Covariates were examined by log–log plot to determine their ability to be incorporated in multivariate models, and tests for interactions were performed. For continuous variables, the proportional hazard assumptions were also assessed by studying the graphs of Schoenfelds residuals.
Results
In total, 55 (12%) of the breast cancer cases showed strong QSOX1 staining, whereas 403 cases showed low expression. Images of tumors with high and low QSOX1 expression are displayed in Figure 1.
Association of QSOX1 Expression with Clinico-Pathologic and Immunohistochemical Features
High QSOX1 expression was associated with high histologic grade, ER and PR negativity, HER2 positivity, and P-cadherin positivity, as summarized in Table 1. No association was found between QSOX1 expression and axillary lymph node status.
QSOX1 Expression in Relation to Proliferation
High QSOX1 expression showed significant associations with increased tumor cell proliferation assessed by Ki67 and mitotic count. (Table 1, see also Figure 2).
QSOX1 Expression in Molecular Subgroups
There were significant differences in QSOX1 expression according to molecular subgroups (Kruskal–Wallis H-test, P<0.001). In the triple-negative subgroup, 14 out of 46 cases (30%) showed high QSOX1 expression, (Table 2), whereas in the HER2+ subgroup, 8 out of 23 cases (35%) showed high QSOX1 level. A subset of the lumB/HER2+ cases (13 cases/32%) also showed high QSOX1 expression.
QSOX1 Expression in Relation to Patient Outcome
Within the QSOX1 low group, no differences in survival were demonstrated for cases showing negative staining (staining index 0–1) compared with cases with weak to moderate staining (staining index 2–6). These subgroups were therefore merged (QSOX1 low). High QSOX1 expression was significantly associated with reduced breast cancer-specific survival in univariate survival analysis (Figure 3, log-rank test, P<0.001) with 10 year survival of 67% for cases showing high QSOX1 expression compared to 89% for cases with low expression (Table 3). Univariate Cox survival analysis showed poor outcome for cases with high QSOX1 expression with a hazard ratio of 3.07 (confidence interval 1.83–5.16, P<0.001).
In the HER2+/HR− subgroup (n=23) high QSOX1 expression was a significant prognosticator in univariate survival analysis (Supplementary Figure 1A, log-rank test, P=0.003). In the triple-negative subgroup (n=46), high QSOX1 levels were significantly associated with reduced survival (Supplementary Figure 1B, log-rank test, P=0.004). No associations were demonstrated between QSOX1 expression and outcome within the luminal/HER2-negative category (data not shown).
Subsequently, we performed multivariate Cox survival analyses including the basic prognostic factors age, tumor size, histologic grade, and lymph node status in addition to QSOX1 expression. In this model, tumor size, nodal status, and high QSOX1 expression were independent predictors of prognosis (Table 4). By adding the luminal status to the model (luminal versus non-luminal); tumor size, nodal status, QSOX1, and luminal status retained independent prognostic significance (data not shown). There was no interaction between QSOX1 and luminal status. Finally, an extended model with additional adjustment for CK5/6 and P-cadherin was examined. Here, independent prognostic impact was shown for tumor size, nodal status, and QSOX1 expression (data not shown).
Discussion
Here, we examined the QSOX1 protein expression in a large, population-based series of breast carcinomas with long and complete follow-up. Our findings indicate that high QSOX1 expression is associated with an aggressive tumor phenotype characterized by high histologic grade, increased proliferative rate, hormone receptor negativity, HER2 positivity, and reduced survival in both univariate and multivariate models. Consistent with this, elevated QSOX1 mRNA and protein levels in breast cancer have previously been related to high histologic grade.9, 11 In line with our results, an association between high QSOX1 protein expression and increased Ki67 has been shown.11 Moreover, knockdown of QSOX1 in an animal model led to reduced tumor growth, suggesting that QSOX1 promotes proliferation in vivo.23 In contrast, high QSOX1 expression has been correlated with decreased proliferation in vitro24 and reduced tumor growth in vivo.13
We included the basal markers CK5/6 and P-cadherin as a refinement of the triple-negative subgroup.15, 17 High QSOX1 levels were associated with increased P-cadherin expression, whereas the association with CK5/6 did not reach statistical significance (P=0.074).
A previous study showed higher levels of QSOX1 mRNA in ER-negative breast cancer cell lines compared to ER-positive cells.25 One study reported that QSOX1 was repressed by estrogen stimulation in an ER-positive breast cancer cell line.26 Contrasting this, an association between high QSOX1 and ER positivity was suggested by Katchman et al.,11 although this was not confirmed in our study.
To the best of our knowledge, this is the first report of QSOX1 protein expression and prognosis in breast cancer. In our study, high QSOX1 level was associated with reduced survival in univariate analysis, and also showed independent prognostic impact in multivariate Cox analyses. This is in line with a previous report where high QSOX1 gene expression was associated with poor prognosis,11 although we could not confirm that the prognostic impact was restricted to the Luminal B subgroup. We found no interaction between QSOX1 expression and luminal status, and in subgroup analyses, QSOX1 was prognostic within the HER2 positive and triple-negative subgroups.
In contrast, high QSOX1 mRNA expression has been associated with improved prognosis,13 and increased QSOX1 expression has been suggested to protect breast cancer cells against apoptosis,24 and to inhibit autophagy in vitro.27 Contrasting to this, Katchman et al.11 found no correlation between QSOX1, apoptosis, or autophagy.
High QSOX1 expression in breast, pancreatic and renal cancer cell lines has been related to increased invasiveness.11, 12, 23 Also, an association between high QSOX1 and post-translational activation of MMP-2 and MMP-9 has been suggested.12 In breast cancer, MMP-2 and MMP-9 are upregulated in response to HIF-1 expression, and they facilitate invasion through degradation of collagen.28
Intriguingly, a recent report indicated that QSOX1 expression in pancreatic cancer cells were induced by HIF-1 in vitro, with concurrent increased invasiveness.29 Two binding sites for HIF response elements were identified in the promoter region of the QSOX1 gene, implicating that QSOX1 is a target gene for HIF-1. QSOX1 silencing led to decreased invasiveness. Moreover, it has been demonstrated that QSOX1 secreted from fibroblasts is required for proper laminin incorporation into the extracellular matrix, thus supporting the migration of lung cancer cells in vitro.2
Interestingly, QSOX1 might represent a possible therapeutic target. QSOX1 antibodies have been developed,2, 30 and QSOX1 inhibitors have been assessed both in vitro and in vivo.23 QSOX1 might therefore represent a novel and important biomarker in breast cancer. However, it should be mentioned that in the current study, outcome was assessed within a context where the treatment administered may not have been homogeneous. The lack of treatment information on a case level may represent a limitation in prognostic analyses.
Conclusion
Taken together, our findings indicate that high QSOX1 protein expression reflects adverse tumor features and reduced breast cancer survival. The function of QSOX1 in these tumors is presently not clear and further studies are needed. Still, QSOX1 expression may represent both a biomarker for aggressive disease and a future treatment target.
References
Chakravarthi S, Jessop CE et al, Willer M, Intracellular catalysis of disulfide bond formation by the human sulfhydryl oxidase, QSOX1. Biochem J 2007;404:403–411.
Ilani T, Alon A, Grossman I et al, A secreted disulfide catalyst controls extracellular matrix composition and function. Science 2013;341:74–76.
Thorpe C, Hoober KL, Raje S et al, Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch Biochem Biophys 2002;405:1–12.
Coppock DL, Thorpe C . Multidomain flavin-dependent sulfhydryl oxidases. Antioxid Redox Signal 2006;8:300–311.
Coppock DL, Kopman C, Scandalis S et al, Preferential gene expression in quiescent human lung fibroblasts. Cell Growth Differ 1993;4:483–493.
Lake DF, Faigel DO . The emerging role of QSOX1 in cancer. Antioxid Redox Signal 2014;21:485–496.
Ouyang X, DeWeese TL, Nelson WG et al, Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res 2005;65:6773–6779.
Song H, Zhang B, Watson MA et al, Loss of Nkx3.1 leads to the activation of discrete downstream target genes during prostate tumorigenesis. Oncogene 2009;28:3307–3319.
Soloviev M, Esteves MP, Amiri F et al, Elevated transcription of the gene QSOX1 encoding quiescin Q6 sulfhydryl oxidase 1 in breast cancer. PLoS One 2013;8:e57327.
Antwi K, Hostetter G, Demeure MJ et al, Analysis of the plasma peptidome from pancreas cancer patients connects a peptide in plasma to overexpression of the parent protein in tumors. J Proteome Res 2009;8:4722–4731.
Katchman BA, Ocal IT, Cunliffe HE et al, Expression of quiescin sulfhydryl oxidase 1 is associated with a highly invasive phenotype and correlates with a poor prognosis in Luminal B breast cancer. Breast Cancer Res 2013;15:R28.
Katchman BA, Antwi K, Hostetter G et al, Quiescin sulfhydryl oxidase 1 promotes invasion of pancreatic tumor cells mediated by matrix metalloproteinases. Mol Cancer Res 2011;9:1621–1631.
Pernodet N, Hermetet F, Adami P et al, High expression of QSOX1 reduces tumorogenesis, and is associated with a better outcome for breast cancer patients. Breast Cancer Res 2012;14:R136.
Knutsvik G, Stefansson IM, Aziz S et al, Evaluation of Ki67 expression across distinct categories of breast cancer specimens: a population-based study of matched surgical specimens, core needle biopsies and tissue microarrays. PLoS One 2014;9:e112121.
Collett K, Stefansson IM, Eide J et al, A basal epithelial phenotype is more frequent in interval breast cancers compared with screen detected tumors. Cancer Epidemiol Biomarkers Prev 2005;14:1108–1112.
Nalwoga H, Arnes JB, Wabinga H et al, Expression of aldehyde dehydrogenase 1 (ALDH1) is associated with basal-like markers and features of aggressive tumours in African breast cancer. Br J Cancer 2010;102:369–375.
Arnes JB, Brunet JS, Stefansson I et al, Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res 2005;11:4003–4011.
Aas T, Borresen AL, Geisler S et al, Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 1996;2:811–814.
Straume O, Akslen LA . Alterations and prognostic significance of p16 and p53 protein expression in subgroups of cutaneous melanoma. Int J Cancer 1997;74:535–539.
Foulkes WD, Stefansson IM, Chappuis PO et al, Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 2003;95:1482–1485.
Goldhirsch A, Winer EP, Coates AS et al, Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013 24:2206–2223.
Goldhirsch A, Wood WC, Coates AS et al, Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011 22:1736–1747.
Hanavan PD, Borges CR, Katchman BA et al, Ebselen inhibits QSOX1 enzymatic activity and suppresses invasion of pancreatic and renal cancer cell lines. Oncotarget 2015;6:18418–18428.
Morel C, Adami P, Musard JF et al, Involvement of sulfhydryl oxidase QSOX1 in the protection of cells against oxidative stress-induced apoptosis. Exp Cell Res 2007;313:3971–3982.
Coppock D, Kopman C, Gudas J et al, Regulation of the quiescence-induced genes: quiescin Q6, decorin, and ribosomal protein S29. Biochem Biophys Res Commun 2000;269:604–610.
Inoue A, Yoshida N, Omoto Y et al, Development of cDNA microarray for expression profiling of estrogen-responsive genes. J Mol Endocrinol 2002;29:175–192.
Poillet L, Pernodet N, Boyer-Guittaut M et al, QSOX1 inhibits autophagic flux in breast cancer cells. PLoS One 2014;9:e86641.
Gilkes DM, Semenza GL, Wirtz D . Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer 2014;14:430–439.
Shi CY, Fan Y, Liu B et al, HIF1 contributes to hypoxia-induced pancreatic cancer cells invasion via promoting QSOX1 expression. Cell Physiol Biochem 2013;32:561–568.
Grossman I, Alon A, Ilani T et al, An inhibitory antibody blocks the first step in the dithiol/disulfide relay mechanism of the enzyme QSOX1. J Mol Biol 2013;425:4366–4378.
Acknowledgements
We thank Gerd Lillian Hallseth, Randi Hope Lavik and Bendik Nordanger for excellent technical assistance. This work was partly supported by the Research Council of Norway through its Centres of Excellence funding scheme, project number 223250. GK is supported by grant No 911637 from the Helse Vest Research Fund. LAA is supported by other grants from the Norwegian Cancer Society, the Research Council of Norway and Helse Vest Research Fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Knutsvik, G., Collett, K., Arnes, J. et al. QSOX1 expression is associated with aggressive tumor features and reduced survival in breast carcinomas. Mod Pathol 29, 1485–1491 (2016). https://doi.org/10.1038/modpathol.2016.148
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.148
This article is cited by
-
Effects of redox modulation on quiescin/sulfhydryl oxidase activity of melanoma cells
Molecular and Cellular Biochemistry (2023)
-
Obesity-associated methylation in breast tumors: a possible link to disparate outcomes?
Breast Cancer Research and Treatment (2020)
-
Core fucosylated glycan-dependent inhibitory effect of QSOX1-S on invasion and metastasis of hepatocellular carcinoma
Cell Death Discovery (2019)