Abstract
Several high-risk human papillomavirus (HPV)-induced cell biomarkers have been proposed as possible candidates to identify patients harboring high-grade squamous intraepithelial lesions (HSILs) of the uterine cervix. We aimed to determine the feasibility of the detection of the mRNA of six biomarkers in cervical smear specimens obtained by liquid-based cytology and to evaluate whether this approach might be useful in the identification of patients with HSIL. One-hundred and twenty three women referred to colposcopy in the Hospital Clinic of Barcelona were included in the study. After a thorough study, including Pap test, high-risk HPV testing (Hybrid Capture 2 test), and colposcopy with directed biopsy and/or endocervical curettage, 48 patients were diagnosed with HSIL, whereas 75 were classified as negative (n=28), or harboring low-grade SIL (n=47). CDKN2A/p16, BIRC5, MMP9, TOP2A, MCM5, and MKI67 mRNA expression was analyzed by reverse transcription quantitative polymerase chain reaction in liquid-based cytology after the Pap test and Hybrid Capture 2 performance. The tissue expression of these biomarkers was analyzed by immunohistochemistry in the biopsy material. One-hundred and thirteen out of 123 (92%) liquid-based cytology yielded adequate material for mRNA analysis. TOP2A was the most sensitive (97%) biomarker for the detection of HSIL and CDKN2A/p16 the most specific (78%). The combination of TOP2A and CDKN2A/p16 showed a sensitivity of 96% (95% confidence interval (CI): 88–99) and a specificity of 71% (95% CI: 55–82). In the immunohistochemistry analysis, all biomarkers showed a high sensitivity but low specificity for HSIL, except CDKN2A/p16 which had a sensitivity of 100% and a specificity of 63%. The combination of TOP2A and CDKN2A/p16 showed a sensitivity of 100% (95% CI: 91–100) and a specificity of 43% (95% CI: 32–55). The detection of mRNA of cell biomarkers in liquid-based cytology material is feasible. The combination TOP2A and CDKN2A/p16 has a good balance between sensitivity and specificity for the detection of women with HSIL.
Similar content being viewed by others
Main
Screening programs based on cervical cytology (Pap test) have led to a decrease in the incidence and the mortality by cervical cancer. However, the efficacy of cytological screening is hampered by the suboptimal sensitivity and interobserver variability of the conventional Pap test.1, 2 Although the introduction of liquid-based cytology has not led to improvements in terms of relative sensitivity for the detection of cervical cancer precursors,3 this technique has enabled the use of adjunctive tests that may help to reduce the rate of false-negative cytological results and add objectivity to the classical morphological evaluation.
High-risk human papillomaviruses (HPV) are the causative agents of cervical cancer, and high-risk HPV testing has shown to improve the sensitivity of the Pap test to over 95%.4, 5 However, high-risk HPV tests have a lower specificity than the Pap test, and positive results are frequently observed in women without cervical lesions or who show mild abnormalities, such as atypical squamous cells of unknown significance (ASC-US) or low-grade squamous intraepithelial lesions (LSILs). In most women with these low-risk lesions, both high-risk HPV infection and the epithelial abnormalities are cleared within a few months or years (transient infections). Therefore, there is a need to explore new more specific techniques that may help to identify the subset of women harboring high-grade squamous intraepithelial lesions (HSIL),6 the true precursor of CC, from the broad group of women testing positive for a high-risk HPV.
In recent years, several high-risk HPV-induced cellular molecules involved in replication, transcription, DNA repair, apoptosis, proliferation, and invasion and metastasis have been analyzed as possible biomarkers to detect women at risk of cervical cancer development. The list of candidates who have shown potential utility in cervical cancer screening includes p16INK4a (CDKN2A), survivin (BIRC5), metalloproteinase 9 (MMP9), topoisomerase 2 alpha (TOP2A), minichromosome maintenance 5 (MCM5), or MKi67 proteins (MKI67).7, 8, 9 Most studies focused on these biomarkers have used immunohistochemistry to detect protein expression. However, several shortcomings of immunohistochemistry, in particular the interobserver variation inherent to all morphological techniques and the difficulty to obtain reproducible quantification, hamper proper evaluation of the clinical usefulness of these biomarkers. mRNA detection has the advantage of being less subjective than morphologic assessment. However, very little data are available on the use of mRNA-based techniques to detect these cell biomarkers in liquid-based cytology cervical samples, and their possible value in cervical cancer prevention has not been proven.
In the present study we aimed to determine the feasibility of the detection of the mRNA of six biomarkers in liquid-based cytology preserved samples and to evaluate whether this approach might be useful in the identification of patients with HSIL.
Materials and methods
Study Design and Case Selection
This prospective study included 123 women referred to the colposcopy clinic of the Department of Obstetrics and Gynecology of the Hospital Clinic of Barcelona from January to December 2010. Referral to colposcopy was based on a positive result in high-risk HPV testing or a Pap test result of ASC, atypical glandular cells, LSIL, HSIL, or cervical cancer within the 6 months previous to admission. At the initial visit in the colposcopy clinic, all women underwent a Pap test, high-risk HPV testing, a colposcopy examination, and at least one biopsy for histological analysis (colposcopically directed biopsy and/or endocervical curettage).
We selected women who fulfilled the following inclusion criteria: (1) adequate cervical cytology, and (2) colposcopically directed biopsy and/or endocervical curettage, (3) high-risk HPV testing taken concurrently, and (4) concordant results in the cytological, histological and virological tests. Concordant results included in the study were in the following three diagnostic categories: (a) negative: women with a negative Pap test result for intraepithelial lesion or carcinoma, a negative result in biopsy and negative high-risk HPV testing (n=28); (b) LSIL: women with a Pap test result of LSIL, a biopsy showing LSIL and a positive high-risk HPV testing (n=47); and (c) HSIL: patients with Pap test result of HSIL, biopsy confirming HSIL and positive high-risk HPV testing (n=48).6
The exclusion criteria were as follows: (1) histological diagnosis of carcinoma or previous history of CC; (2) previous treatment for CIN2-3 performed within the previous 3 years; (4) pregnancy; (5) previous HPV vaccine, and (6) discordant results in cytology, histology, and Hybrid Capture 2 testing (eg patients with Pap test result of HSIL and biopsy with LSIL or negative result or women with positive high-risk HPV and normal cytology).
The study was approved by the institutional ethical review board. All women had signed informed consent.
Patient Management
Prior to the colposcopy procedure, a cervical sample was collected in all women using a cytobrush, which was transferred to PreservCyt solution (Hologic , Marlborough, MA, USA). The first part of the sample was used for ThinPrep liquid-based cytology. The residual material was used first for high-risk HPV testing and second RNA isolation.
Colposcopy was performed using a Olympus Evis Exera II CV-180 colposcope (Olympus, Barcelona, Spain) after preparing the cervix with 5% acetic acid. A colposcopically directed biopsy was taken in all patients on the identificaton of an abnormal area.10 When the transformation zone was not completely visible, an endocervical curettage using a Kervokian curette was also performed. A random biopsy from the transformation zone was performed in all the women with a completely visible transformation zone having no colposcopic abnormalities.
Liquid-based Cytology, Histological Diagnosis
Thin-layer cytology slides were prepared using the Thinprep T2000 slide processor (Hologic) and stained using the Papanicolaou method. Cytology slides were evaluated by a cytotechnologist and confirmed by a pathologist using the revised Bethesda nomenclature.11 Subsequently, liquid-based cytology samples were centrifuged and the pellets stored at −80 °C until processing.
Formalin-fixed, paraffin-embedded 4-μm sections were routinely stained with hematoxylin and eosin (H&E). All the histological samples were reviewed by one of the authors (JO) to confirm the presence or absence of SIL and its grade. The histological diagnoses were established using pure morphologic criteria based on the H&E-stained sections, with no knowledge of HPV status or the cytology result.
High-risk HPV detection (types 16, 18, 32, 34, 36, 39, 45, 51, 52, 56, 58, 59, and 68) was performed using the Hybrid Capture 2 System (Qiagen, Gaithersburg, MD, USA) on the material collected in liquid-based media (PreservCyt). A relative light unit of 1 (1.0 pg/ml) was used as the cut-off to classify a specimen as positive for high-risk HPV.12
RNA Isolation, Reverse Transcriptase Polymerase Chain Reaction and Quantitative PCR
Once cytologic diagnosis and high-risk HPV testing were performed, 5 ml of each cell suspension was centrifuged at 2200 g for 15 min, the cell pellet was mixed with 700 μl QIAzol lysis reagent and stored at −70 °C (Qiagen, Hilden, Germany). Total RNA was extracted using the RNeasy RNA extraction kit (Qiagen), according to the manufacturer’s protocol. RNA concentrations were measured with a NanoDrop instrument (Thermo Scientific, Wilmington, DE, USA). Samples with an RNA yield lower than 1.5 μg were excluded from the analysis.
Reverse transcription was performed starting with 10 μl of total RNA in a 20-μl reaction volume using random hexamers and the High Capacity cDNA RT-kit from Applied Biosystems (Foster City, CA, USA) according to the manufacturer’s protocol. A reaction without RT was run in parallel for each specimen to control for DNA contamination.
All polymerase chain reactions (PCRs) were performed in triplicate at a reaction volume of 25 μl containing 5 μl of cDNA, diluted 1:10, mixed with Taqman Universal PCR MasterMix (Applied Biosystems). The following protocol was used for all assays: denaturation (95 °C for 10 min) and amplification (95 °C for 20 s, 60 °C for 1 min) repeated for 40 cycles.
The expression of CDKN2A/p16, BIRC5, MMP9, TOP2A, MCM5, and MKI67 was analyzed. Beta glucuronidase (GUSB) and cGMP-dependent protein kinase 1 (PKG1) were selected as reference genes for normalization. This combination of reference genes demonstrated a high stability in expression between groups of normal samples versus HSIL samples. For stability comparisons of candidate reference genes, the GeNorm and Normfinder programs were used, which are included as applications in the GenEx software (MultiD analysis, Sweden).
The number of cycles required for the signal to cross the threshold (cycle threshold value) for target genes was set at 40 cycles. Cycle threshold levels are inversely proportional to the amount of target nucleic acid in the sample (the lower the cycle threshold level, the greater the amount of target nucleic acid in the sample). For the reference gene PRKG1, a cycle threshold value above 32 cycles indicates poor RNA quality. Samples above this cycle threshold values were therefore considered invalid and excluded from the analysis.
Primers and probes sequence used in the quantitative PCR (q-PCR) are listed in Table 1. All probes were FAM-labeled and all reactions were run in singleplex.
Immunohistochemistry
Immunohistochemistry was performed in the simultaneous biopsy of the patients included in the study with the automated system Autostainer Link 48 (Dako, Carpinteria, CA, USA) using the EnVision system (Dako). The primary antibodies, the dilutions, and the sources are listed in Table 1.
Each series included a positive control consisting of a HSIL of the uterine cervix. In all cases, the evaluation of the positivity was restricted to the area showing the lesion. Staining for all antibodies was scored as negative, focal, or diffuse on the basis of nuclear and/or cytoplasmic staining. Staining or reactivity in rare isolated cells (<5%) was considered as negative. Cases with more than 5% of positive cells were evaluated as positive (5–25% of positive cells evaluated as focal positivity, and cases with more than 25% of positive cells as diffuse staining). For CDK2A/p16, the positivity was scored as focal for cases with either focal staining of basal cells or any type of staining in suprabasal layers, and diffuse when all basal and suprabasal cells in an area stained positive.13 The histoscore method was also used to further evaluate the immunohistochemistry expression of the biomarkers.14 This semiquantitative immunohistochemical method consists of a sum of the percentages of positively stained cells multiplied by a weighted intensity of staining: Histoscore 1/4 SPi(i+1), in which Pi is the percentage of stained cells in each intensity category (0–100%), and i is the intensity indicating weak (i 1/4 1), moderate (i 1/4 2), or strong staining.
Data Analysis
The data were analyzed with the program SPSS (Version 18.0). The χ or the Fisher’s exact test was used for comparisons between categorical variables as appropriate. The ANOVA test was used to compare quantitative variables between the different categories. Correlations and differences between mRNA expression and immunohistochemistry categories and histoscore were assessed using the Spearman’s rank correlation coefficient (rs). The efficacy of mRNA expression and immunohistochemistry of the biomarkers for the diagnosis of HSIL was evaluated as sensitivity and specificity. All data related to expression are presented as ΔCycle threshold (cycle thresholdtarget−cycle thresholdreference). The level of significance between target gene expression in normal samples and tumor samples was analyzed using ROC analysis.
Results
The total RNA yield varied between 0.5 and 64.3 μg, reflecting variability of cell numbers in the specimens. One-hundred thirteen out of 123 (92%) samples were considered as adequate and only 9% (11/123) were excluded due to low expression of the reference genes and/or low RNA yield. Within the samples excluded 1 was classified as negative, 2 were classified as LSIL, and 7 as HSIL. Thus, 113 samples were considered as adequate for analysis. Of these samples 27/113 (24%) were classified as negative, 45/113 (40%) as LSIL, and 41/113 (36%) as HSIL.
Table 2 shows the mean normalized ratios for the different biomarkers according to the diagnostic group. High ΔCycle threshold values correspond to low expression of a marker, whereas a low ΔCycle threshold corresponds to a high expression of that marker. CDKN2A/p16 expression was significantly higher in HSIL compared with negative specimens (P<0.001). BIRC5 expression was significantly higher in HSIL compared with LSIL specimens (P=0.007) and negative specimens (P<0.001). MMP9 expression was significantly higher in HSIL compared with LSIL samples (P=0.016) and with negative samples (P=0.016). TOP2A and MKI67 expression was significantly higher in HSIL compared with LSIL (P<0.001) or negative samples (P<0.001).
The ROC curves and the area under curve for the identification of patients with HSIL for the different biomarkers are shown in Figure 1. Table 3 shows the sensitivity and specificity of mRNA expression of the different biomarkers for the identification of patients with HSIL after applying the best cut-off values obtained after analysis of the ROC curves. TOP2A was the most sensitive biomarker and CDKN2A/p16 the most specific. The combination of TOP2A and CDKN2A/p16 showed a sensitivity of 96% (95% confidence interval (CI): 88–99) and a specificity of 71% (95% CI: 55–82). The ROC curve and the area under curve for the identification of patients with HSIL for the TOP2A and CDKN2A/p16 combination is shown in Figure 2. Other combinations of biomarkers did not provide any additional value (data not shown).
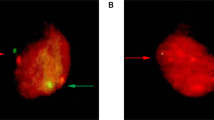
Immunohistochemistry staining for the different biomarkers analyzed was positive in most of the HSIL biopsies. Table 4 shows the sensitivity and specificity of immunohistochemistry of the different biomarkers for the identification of patients with HSIL. All biomarkers, except CDKN2A/p16, showed a high sensitivity but low specificity values. When immunohistochemistry positivity was restricted only to cases showing diffuse staining (over 25% of positive cells), the specificity for all the biomarkers significantly increased, but the sensitivity dropped to low values. The immunostaining for all the biomarkers included in the analysis in a biopsy of a HSIL lesion is shown in Figure 3.
There was no correlation between ΔCycle threshold values and immunohistochemistry staining categories for BIRC5 (rs=−0.072; P=0.533), TOP2A (rs=−0.103; P=0.371), MCM5 (rs=−0.134; P=0.247), and MKI67 (rs=−0.055; P=0.652). No adequate staining was obtained for MMP9. A significant correlation was found between ΔCycle threshold values and immunohistochemistry staining for CDKN2A/p16 (rs=−0.338; P<0.001). The correlation between ΔCycle threshold values and the immunohistochemistry histoscore was not statistically significant for any of the biomarkers analyzed (data not shown).
Discussion
In recent years, the molecular events associated with cervical carcinogenesis have been unraveled, thereby allowing the identification of new molecules that might be potentially useful as biomarkers for cervical cancer prevention. Analysis of the expression of the host-cell genes involved in cervical carcinogenesis may enhance the ability of HSIL detection, serving as a better risk evaluation factor than merely for HPV-DNA detection.
This study shows that the detection of the mRNA expression of different cell biomarkers in liquid PAP specimens from cervical samples is feasible. Thus, molecular assays designed to detect these markers in specimens preserved in liquid-based cytology media may be useful for HSIL detection. In our study almost all biomarkers showed sensitivities higher than 81% for HSIL. TOP2A, an enzyme that unknots and decatenates DNA for DNA replication and cell cycle progression, demonstrated the best sensitivity (96%), being much higher than that of cytology and similar to that shown by high-risk HPV tests.12, 15 CDKN2A/p16 had the lowest sensitivity for HSIL diagnoses but had the highest specificity (69%) compared with other biomarkers. The combination of TOP2A and CDKN2A/p16 resulted in an adequate balance between sensitivity and specificity, and could be used in the identification of HSIL.
Most previous studies have assessed these biomarkers by immunohistochemistry. There is extensive evidence showing that CDKN2A/p16 staining in histological sections increases the detection of HSIL13, 16 and can identify LSIL at higher risk of progression.10, 17 Recently, the combination of p16 and Ki67 detected by immunohistochemistry has been used in cytological samples to identify patients with high-grade cervical lesions.18, 19 In our study, the analysis of these biomarkers in biopsy material by immunohistochemistry showed that all were positive in a high proportion of the HSIL lesions, which is consistent with current evidence.8, 16, 20, 21, 22, 23 Nevertheless, immunohistochemistry is not adequate for screening purposes. Finally, immunohistochemistry only analyzes a small area of the lesion, which may not be representative of the whole process that could explain the absence of correlation between the mRNA expression in the liquid-based cytology and the immunohistochemistry staining in the simultaneous biopsy for most of the biomarkers. Indeed, the main advantages of mRNA detection using q-PCR in liquid-based cytology are that it is less subjective than morphologic assessment and it allows evaluation of the whole cervical mucosa.24
Extraction of high-quality RNA is a prerequisite for the application of RNA transcripts as biomarkers for clinical purposes.25 The use of mRNA from cervical scrapes, which consist of mucus and exfoliated cervical cells, may be challenging because the RNA might be degraded in this material due to their lability. In our study, 91% of the specimens had RNA of adequate quality and could be analyzed. Storage solutions and sample processing are critical steps to ensure high-quality RNA. We centrifuged liquid-based cytology samples remaining after conventional diagnostic tests, and the pellets were stored at −80 °C until processing. The good results obtained indicate that this procedure has a relatively minor effect on the quality of mRNA. Moreover, in the present study, mRNA extraction was performed after cytological diagnosis and high-risk HPV testing by Hybrid Capture 2, and both methods require an important amount of cells. Other screening strategies using smaller quantities of liquid-based cytology might facilitate the extraction of high-quality RNA.
Few studies have addressed the analysis of the mRNA expression of the host-cell genes involved in cervical carcinogenesis. In a previous study by Boulet et al20 CDKN2A/p16 expression did not allow discrimination between women with or without SIL. Nevertheless, in their study the samples were preserved in SurePath media, which seems to result in highly fragmented RNA, not suitable for reverse transcriptase polymerase chain reaction (RT-PCR) assays.26, 27 Contrarily, fixation in PreservCyt has been proven to provide material suitable for RNA isolation and subsequent analysis, as shown in the present study.28, 29 Recently, Bourmenskaya et al30 have shown that a gradual increase in the expression of proliferation markers and a decrease in the expression of pro-apoptotic human genes, as determined by RT-PCR, were associated with increased risks of carcinogenesis. These authors proposed a quantitative model combining the expression of CDKN2A/p16 and BIRC5, which are associated with adverse prognosis,31, 32 and MKI67, which distinguish SIL from normal or benign reactive cervical lesions,22 for cervical cancer risk assessment. Our results also indicate that a combination of biomarkers could be an adequate strategy for HSIL detection. However, the best combination of biomarkers to achieve the highest sensitivity and specificity remains to be determined. In our study the combination of TOP2A and CDKN2A/p16 showed a sensitivity and specificity for HSIL diagnosis of 96 and 71%, respectively.
This study has some limitations. First, changes in the levels of expression and/or functions of host-cell genes due to the presence of high-risk HPV in samples without cellular dysplasia were not evaluated. Negative cytology samples positive for high-risk HPV were not included in the study as HPV testing is not performed in the primary screening of our clinical setting. Second, the correlation of the mRNA expression by q-PCR from the biomarkers analyzed in histological material was not investigated. We only considered cytological samples as our aim was to study the feasibility of mRNA analysis in a standard clinical setting.
In conclusion, we have shown that mRNA expression in liquid-based Pap specimens is feasible and could be used for HSIL detection. The combination of CDKN2A/p16 and TOP2A has shown an adequate sensitivity and specificity to identify women with HSIL. Further studies including a larger number of patients representing the complete reality of the secondary prevention of cervical cancer are warranted to confirm that biomarker identification using strategies based on mRNA detection might have a role in the secondary prevention of cervical cancer.
References
Fahey MT, Irwig L, Macaskill P . Meta-analysis of Pap test accuracy. Am J Epidemiol 1995;141:680–689
Kyndi M, Frederiksen K, Kruger KS . Cervical cancer incidence in Denmark over six decades (1943-2002). Acta Obstet Gynecol Scand 2006;85:106–111
Siebers AG, Klinkhamer PJ, Grefte JM et al. Comparison of liquid-based cytology with conventional cytology for detection of cervical cancer precursors: a randomized controlled trial. JAMA 2009;302:1757–1764
Naucler P, Ryd W, Tornberg S et al. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N Engl J Med 2007;357:1589–1597
Paraskevaidis E, Arbyn M, Sotiriadis A et al. The role of HPV DNA testing in the follow-up period after treatment for CIN: a systematic review of the literature. Cancer Treat Rev 2004;30:205–211
Darragh TM, Colgan TJ, Thomas CJ et al. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Int J Gynecol Pathol 2013;32:76–115
Barbosa LC, da Silva I, Correa JC, Ribalta JC . Survivin and telomerase expression in the uterine cervix of women with human papillomavirus-induced lesions. Int J Gynecol Cancer 2011;21:15–21
Santin AD, Zhan F, Bignotti E et al. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology 2005;331:269–291
Williams GH, Romanowski P, Morris L et al. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci USA 1998;95:14932–14937
del Pino M, Garcia S, Fuste V et al. Value of p16(INK4a) as a marker of progression/regression in cervical intraepithelial neoplasia grade 1. Am J Obstet Gynecol 2009;201:488–7
Solomon D, Davey D, Kurman R et al. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA 2002;287:2114–2119
Ordi J, Alonso I, Torne A et al. Human papillomavirus load in Hybrid Capture II assay: does increasing the cutoff improve the test?. Gynecol Oncol 2005;99:313–319
Bergeron C, Ordi J, Schmidt D et al. Conjunctive p16INK4a testing significantly increases accuracy in diagnosing high-grade cervical intraepithelial neoplasia. Am J Clin Pathol 2010;133:395–406
Casals G, Ordi J, Creus M et al. Expression pattern of osteopontin and alphavbeta3 integrin during the implantation window in infertile patients with early stages of endometriosis. Hum Reprod 2012;27:805–813
zur Hausen H . Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002;2:342–350
Ordi J, Garcia S, del Pino M et al. p16 INK4a immunostaining identifies occult CIN lesions in HPV-positive women. Int J Gynecol Pathol 2009;28:90–97
Hariri J, Oster A . The negative predictive value of p16INK4a to assess the outcome of cervical intraepithelial neoplasia 1 in the uterine cervix. Int J Gynecol Pathol 2007;26:223–228
Ikenberg H, Bergeron C, Schmidt D et al. Screening for cervical cancer precursors With p16/Ki-67 Dual-Stained Cytology: Results of the PALMS Study. J Natl Cancer Inst 2013;105:1550–1557
Ordi J, Sagasta A, Munmany M et al. Usefulness of p16/Ki67 immunostaining in the triage of women referred to Colposcopy. Cancer Cytopathol 2014;122:227–235
Boulet GA, Horvath CA, Depuydt CE, Bogers JJ . Biomarkers in cervical screening: quantitative reverse transcriptase PCR analysis of P16INK4a expression. Eur J Cancer Prev 2010;19:35–41
Branca M, Giorgi C, Santini D et al. Survivin as a marker of cervical intraepithelial neoplasia and high-risk human papillomavirus and a predictor of virus clearance and prognosis in cervical cancer. Am J Clin Pathol 2005;124:113–121
Gupta N, Srinivasan R, Rajwanshi A . Functional biomarkers in cervical precancer: an overview. Diagn Cytopathol 2010;38:618–623
Shroyer KR, Homer P, Heinz D, Singh M . Validation of a novel immunocytochemical assay for topoisomerase II-alpha and minichromosome maintenance protein 2 expression in cervical cytology. Cancer 2006;108:324–330
Tsoumpou I, Arbyn M, Kyrgiou M et al. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev 2009;35:210–220
Cuschieri KS, Beattie G, Hassan S et al. Assessment of human papillomavirus mRNA detection over time in cervical specimens collected in liquid based cytology medium. J Virol Methods 2005;124:211–215
Horvath CA, Boulet G, Sahebali S et al. Effects of fixation on RNA integrity in a liquid-based cervical cytology setting. J Clin Pathol 2008;61:132–137
Powell N, Smith K, Fiander A . Recovery of human papillomavirus nucleic acids from liquid-based cytology media. J Virol Methods 2006;137:58–62
Habis AH, Vernon SD, Lee DR et al. Molecular quality of exfoliated cervical cells: implications for molecular epidemiology and biomarker discovery. Cancer Epidemiol Biomarkers Prev 2004;13:492–496
Martin CM, Astbury K, McEvoy L et al. Gene expression profiling in cervical cancer: identification of novel markers for disease diagnosis and therapy. Methods Mol Biol 2009;511:333–359
Bourmenskaya O, Shubina E, Trofimov D et al. Host gene expression profiling of cervical smear is eligible for cancer risk evaluation. J Clin Pathol 2013;66:282–285
Altieri DC . Survivin in apoptosis control and cell cycle regulation in cancer. Prog Cell Cycle Res 2003;5:447–452
Li F . Survivin study: what is the next wave?. J Cell Physiol 2003;197:8–29
Acknowledgements
We thank Roser Esteve, Silvia Alòs, Francisco Manuel Perez, and Naiara Vega for their help with the evaluation of the Pap tests and Marta Sanchez for her administrative support. We thank Donna Pringle for the English revision of the manuscript. This work was funded by Instituto de Salud Carlos III (ICSiii)-Fondos de Investigación Sanitaria and ERDF ‘one way to Europe’ (PI09/1084, PI09/1524, PI12/01231, and PI12/01165).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
del Pino, M., Svanholm-Barrie, C., Torné, A. et al. mRNA biomarker detection in liquid-based cytology: a new approach in the prevention of cervical cancer. Mod Pathol 28, 312–320 (2015). https://doi.org/10.1038/modpathol.2014.106
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2014.106
This article is cited by
-
TOP2A and CENPF are synergistic master regulators activated in cervical cancer
BMC Medical Genomics (2020)
-
Comprehensive transcriptomic analysis of papillary thyroid cancer: potential biomarkers associated with tumor progression
Journal of Endocrinological Investigation (2020)
-
Correlation between indoleamine 2,3 dioxygenase mRNA and CDKN2A/p16 mRNA: a combined strategy to cervical cancer diagnosis
Medical Oncology (2016)
-
MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer
Tumor Biology (2015)