Abstract
Low-grade serous carcinomas and serous borderline tumors, combined herein and referred to as low-grade serous tumors, show distinct molecular alterations and clinical behaviors compared with high-grade serous carcinomas. The discrimination between low-grade serous tumors and high-grade serous carcinomas can be challenging on small tissue samples, such as cell blocks of paracentesis fluid or biopsies from omental disease. The purpose of this study was to test the ability of TP53 and CDKN2A immunohistochemistry to distinguish between high-grade serous carcinomas and low-grade serous tumors on small tissue samples. Tissue microarrays containing 582 high-grade serous carcinomas, 45 low-grade serous carcinomas, and 49 serous borderline tumors, confirmed by contemporary histopathological review, were stained for TP53 and CDKN2A (DO7 and E6H4 antibody clones, respectively). TP53 was scored as completely absent, wild-type pattern or overexpressed (>60%), and CDKN2A was scored as either negative/patchy (<90%) or block expression (>90%). The combination of the two markers, ie, the TP53 wild-type pattern and CDKN2A patchy expression, had sensitivity for low-grade serous tumors of 89%, a specificity of 93%, a positive predictive value of 68%, and a negative predictive value of 98%. These markers can, therefore, be used on small biopsies/cell blocks to refute a diagnosis of low-grade serous tumors. These findings may inform emerging neoadjuvant therapeutic strategies in advanced ovarian cancers and may be crucial for future clinical trials on molecular-based therapies.
Similar content being viewed by others
Main
Low-grade serous carcinomas are currently not part of the recommended cancer reporting protocols for pathologists, and are, therefore, not incorporated into many synoptic-reporting data structures. However, changes are anticipated as the new edition of the WHO blue book will include low-grade serous carcinomas as a new histological classification. The inclusion of low-grade serous carcinomas as a separate disease entity has taken a long time in part because of the controversy and confusion surrounding the term micropapillary serous carcinoma, and the delineation from serous borderline tumor.1 There is a current consensus that low-grade serous carcinomas requires frank destructive invasion of >5 mm, while serous tumors without frank invasion but with micropapillary architecture should be named serous borderline tumor micropapillary type or noninvasive micropapillary serous carcinoma.2 Low-grade serous carcinomas and serous borderline tumors are associated with mutually exclusive somatic mutations in both KRAS and BRAF genes, which are noticeably absent in the vast majority of high-grade serous carcinomas.3, 4 In contrast, typical high-grade serous carcinomas exhibit ubiquitous TP53 mutations, along with chromosomal rearrangements due to double-stranded DNA repair defects and high rates of proliferation.5, 6, 7
Malpica et al.8, 9, 10 established the histological diagnostic criteria that allowed a reproducible separation of low-grade serous carcinomas from high-grade serous carcinomas. The MD Andersen Cancer Centre group went on to describe the clinical features of low-grade serous carcinomas, which include: presenting in younger patients, frequently in high-stage, and showing lower responses to chemotherapy compared with high-grade serous carcinomas.11 Subsequent studies confirmed that only occasional low-grade serous carcinomas exhibit complete response to standard chemotherapy in the neoadjuvant or recurrent setting.12, 13, 14, 15 With the increasing evidence suggesting that neoadjuvant chemotherapy is an equivalent or superior treatment regimen for advanced stage ovarian cancer, the selection of cases of patients who are most likely to respond remains a foremost clinical problem, which begins with accurate diagnosis. We have recently shown that accurate diagnosis of histological type is possible on whole histological sections and can be further improved by the use of immunohistochemical markers.16, 17 However, central pathology review of the German portion of an international clinical trial has revealed that 3.3% of cases were actually serous borderline tumors overdiagnosed as ovarian carcinoma, and that in 25% of cases, assessment of histological type was inaccurate.18 Given the high degree of intratumoral heterogeneity inherent to high-grade serous carcinomas, differential diagnosis of low-grade serous carcinomas can also be challenging on limited samples in the neoadjuvant setting, where only cell blocks from paracentesis fluid or biopsies of pelvic/peritoneal implants are available. A recent study from Toronto showed that women were treated with neoadjuvant chemotherapy based on clinical diagnosis (CA125/imaging) was in 10% of cases, on cytology-based diagnosis in 78%, and in 12% based on core biopsy.19 The study concluded that ovarian carcinoma diagnosis was confirmed in 96% of cases at interval debulking, but histological type was only accurate in 85% of cases, suggesting difficulties in the assignment of histological type on the initial pretreatment samples.19 Furthermore, in the era of individualized therapy and with the launch of clinical trials evaluating molecular-based targeted therapy, such as GOG239 targeting the MAPK pathway in low-grade serous carcinomas, it is necessary to achieve a more robust diagnosis supported by ancillary techniques on limited biopsy or cytological material.
Differences in the expression of TP53 and CDKN2A between low- and high-grade serous carcinomas were reported previously.20, 21 We have employed this information for the differential diagnosis of low-grade serous tumors (low-grade serous carcinomas and serous borderline tumors) for years, but a formal statistical assessment of the test characteristics has never been published. Therefore, we tested the utility of combined TP53/CDKN2A expression for the distinction between low-grade serous tumors versus high-grade serous carcinomas, using tissue microarrays to simulate the limited samples likely to be available to the pathologist.
Materials and methods
Study Cohorts
For inclusion, cases must have had contemporary pathology review, and a diagnosis of high- and low-grade serous carcinomas, or serous borderline tumors. Low-grade serous carcinomas had to conform to the strict MD Anderson Cancer Center criteria in order to be classified as such.9 The putative site of origin was not used as a case selection criterion; hence, the series included cases with putative ovarian, tubal, or peritoneal origin. Cases must have been successfully assessed for both TP53 and CDKN2A immunohistochemical expression data assessed on a tissue microarray. The cases were pooled from five different sources, including three previously described cohorts: British Columbia moderate risk,22 British Columbia extreme risk,23, 24 and the Vancouver General Hospital tumor bank cohorts.17 The fourth cohort, Tom Baker Cancer Centre cohort, has been previously described.25 Briefly, it represents a selection of 80 advanced stage pelvic (including putative ovarian, peritoneal or fallopian tube origin) serous carcinomas diagnosed and treated at the Tom Baker Cancer Centre, in Calgary, AB from 2003–2007. Forty platinum resistant/intermediate cases (defined by recurrence within 12 months) were matched with 40 platinum sensitive cases (recurrence later than 12 months) for age, residual tumor, stage, neoadjuvant chemotherapy, and number of chemotherapy cycles. All patients received standard surgery with comprehensive staging or debulking, and platinum-containing chemotherapy. Adjuvant chemotherapy was administered within 6 weeks of surgery and considered as completed adequately for all patients. Thirty women out of 80 (38%) received neoadjuvant chemotherapy. Given the rarity of low-grade serous carcinomas, we performed an additional targeted search within the Calgary Laboratory Services Pathology files and the Vancouver General Hosptial tumor bank for additional low-grade serous carcinomas and serous borderline tumors, which formed the fifth cohort, referred to as the Calgary Laboratory Services cohort.
In order to validate the findings on material derived from paracentesis fluid, we retrieved two full sections from cell blocks from paracentesis fluid from 30 unrelated cases (serous borderline tumors, N=10, low-grade serous carcinomas N=10, and high-grade serous carcinomas N=10). Ethics board approval from both institutions was received.
Tissue Microarray Construction and Immunohistochemistry
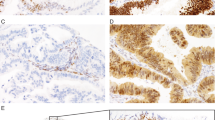
Two new tissue microarrays encompassing the Calgary Laboratory Services and Tom Baker Cancer Centre cohort were built with duplicate 0.6 mm cores (Pathology devices, Westminster, MD, USA). These tissue microarrays and full section from cell blocks were subjected to immunohistochemistry for TP53 and CDKN2A using the Leica Bond platform. For CDKN2A, clone E6H4 (Cin Teq) was used with the company’s pretreatment protocol E2-20 in a dilution of 1:24. For TP53, clone DO7 (DAKO) was used with the pretreatment protocol E2-20 in a dilution of 1:5 000. The other cohorts were previously stained using the same antibodies on the Ventana Benchmark platform.1722, 23, 24 For CDKN2A, the company’s pretreatment protocol was CC1 and the dilution was 1:2. For TP53, the pretreatment protocol was CC1 and the dilution was 1:400. TP53 immunohistochemistry was scored as completely absent, wild-type pattern (between 1and 60% of tumor cell nuclei), or overexpression (>60%). As previously shown, the TP53-staining patterns correlate with the mutational status of TP53 (Figure 1).23, 26 Complete absence of TP53 indicates TP53 null mutation, whereas TP53 overexpression is indicative of TP53 missense mutation, and any staining pattern in between suggests wild-type TP53. The two extreme staining patterns (‘all or nothing’) are combined as aberrant expression, suggesting any type of TP53 mutation. CDKN2A immunohistochemistry was scored as negative/patchy (<90% of tumor cells) and block expression (>90% of tumor cells). Immunohistochemistry for BRAF V600E was performed and evaluated as previously described.27
(a) High-grade serous carcinoma with diffuse TP53 expression. (b) Low-grade serous carcinoma with focal TP53 expression. (c) High-grade serous carcinomas with complete absence of TP53 staining. (d) High-grade serous carcinomas with focal TP53 staining. (e) High-grade serous carcinomas with block CDKN2A staining. (f) Low-grade serous carcinomas with patchy CDKN2A staining, note patched out negative tumor areas.
KRAS Mutation Analysis
One-mm biopsy punches were used to core tissues from formalin-fixed paraffin-embedded blocks using hematoxylin and eosin slides as a guide for maximal tumor retrieval and to prevent non-tumor carryover. DNA was extracted from tissue cores using the QIAamp DNA Micro Kit (Qiagen) according to the manufacturer’s protocol. DNA concentration and purity were determined using a NanoDrop 2000 c spectrophotometer. All samples had 260/280 absorbance values >1.9. KRAS mutational analysis was performed using the KRAS PCR Kit (Qiagen). This ARMS-based assay is capable of detecting seven somatic mutations within codons 12 and 13 of the KRAS gene with 1% sensitivity. The assay was run on an ABI 7500 Fast real-time PCR System under standard conditions, according to the manufacturer’s protocol.
Statistical Analysis
Unsupervised multivariate clustering analysis of TP53 and CDKN2A immunohistochemistry scores was performed using the Ward algorithm. The frequency of low- and high-grade serous carcinomas were tabulated for each of the six resultant clusters. The TP53/CDKN2A diagnostic test performance was quantified with sensitivity and specificity calculations using morphological classification as the gold standard. Sensitivity is defined as the proportion of a diagnosis that is thought to be present and is correctly identified by the test, and specificity is defined as the proportion of a diagnosis that is thought to be absent and is correctly identified by the test. Positive and negative predictive value were calculated in order to determine the precision of the diagnostic test. Positive predictive value is defined as the probability of a positive diagnosis, given that there was a positive test result, whereas negative predictive value is defined as the probability of a negative diagnosis, given that there was a negative test result. Confidence intervals (95%) were calculated for all sensitivity, specificity, and positive and negative predictive value calculations. All statistical analyses were performed by SEK using JMP v 10.0.0 (SAS Institute, Cary, NC, USA).
Results
The background demographics of the study cohorts are depicted in Table 1. The immunohistochemical results for TP53 and CDKN2A across cohorts are shown in Tables 2 and 3 with representative stains in Figure 1. The positivity rate for CDKN2A staining in high-grade serous carcinomas (defined by diffuse block staining in more than 90% of tumor cells) ranged from 55 to 72% across cohorts. Aberrant TP53 staining in high-grade serous carcinomas was seen in 86–93% of cases. This relative consistency in staining pattern allowed us to combine the cohorts for an overall analysis. In addition, because of the similarity of CDKN2A and TP53 expression in serous borderline tumors and low-grade serous carcinomas, we combined both groups to form a new group of low-grade serous tumors.
In order to assess the diagnostic performance of a combination of TP53 with CDKN2A staining, we performed unsupervised hierarchical two-way cluster analysis utilizing Ward’s algorithm. With two markers (TP53 with three possible outcomes and CDKN2A with two outcomes), six combinations/cluster groups were derived (Figure 2). Five of these cluster groups were associated with the histological type diagnosis of high-grade serous carcinomas. Clusters 1 and 3 (CDKN2A block and TP53 overexpression or complete absence), together accounting for almost half of the cases, were all diagnosed as high-grade serous carcinomas by the gold standard morphology. Clusters 4 and 6 (CDKN2A patchy and TP53 overexpression or complete absence), together accounting for 29% of the cases, were in >95% diagnosed as high-grade serous carcinomas. Cluster 2 formed the smallest group with 4% of the cases, still most likely associated with high-grade serous carcinoma. The majority of low-grade serous carcinomas were found in cluster 5 (CDKN2A patchy and TP53 wild-type staining).
Under the assumption that only cluster 5 is predictive for low-grade serous carcinomas and the remaining cluster groups for high-grade serous carcinomas, the sensitivity to predict low-grade serous carcinomas is 89% (95% CI, 81–95%) with a specificity of 93% (95% CI, 91–95%) (Table 4). In order to test the transferability of the results to cell blocks, 30 cases were stained. All 20 low-grade serous tumors (10 serous borderline tumors and 10 low-grade serous carcinomas) showed the staining pattern of cluster 5. All 10 high-grade serous carcinomas showed a staining pattern different from cluster 5, suggesting that the results can be reliably transferred to cell blocks.
Given the restrictive morphological criteria used to diagnose low-grade serous carcinomas, we were concerned on the fact that we possibly under-diagnosed low-grade serous carcinomas. Therefore, we assessed mutations known to occur in low-grade serous carcinomas in a subset of cases. With the availability of a BRAF V600E mutation-specific antibody, tissue microarrays of the low-grade serous carcinomas and Tom Baker Cancer Centre cohorts were stained for the BRAF V600E mutation. Owing to high costs, KRAS mutational testing was only performed on 25 cases from the low-grade serous carcinomas and Tom Baker Cancer Centre cohorts. We randomly selected 9 low- and 13 high-grade serous carcinomas, the latter from the immunophenotypic clusters 2 and 5. The results are shown in Table 5. KRAS mutation was detected in two out of nine low-grade serous carcinomas, but not in any of the high-grade serous carcinoma cases. BRAF mutation testing using the mutation-specific antibody did not yield a single positive low-grade serous carcinomas, whereas 23% of serous borderline tumors were positive for the mutation.
Discussion
Our data provide a statistical validation for the use of combined TP53/CDKN2A immunohistochemistry in the differential diagnosis of low-grade serous tumors from high-grade serous carcinomas on small tissue samples. For the purpose of this study, low-grade serous carcinomas and serous borderline tumors were combined to low-grade serous tumors, because we presumed that differential diagnosis of low-grade serous carcinomas from serous borderline tumors is not possible on ascites fluid derived by paracentesis or on small omental biopsy solely based on morphological grounds. Immunohistochemical staining resulting in a TP53 wild-type pattern and CDKN2A showing patchy or an absence of expression has a sensitivity of 89% and a specificity of 93% for low-grade serous tumors. We believe that the performance of this test presents compelling evidence for the adjunct use of this two stain panel on the differential diagnosis of high-grade serous carcinomas and low-grade serous tumors in difficult cases, such as samples with limited tissue availability. The strength of the test is the negative predictive value (98%), meaning that if certain staining patterns are encountered, eg, aberrant TP53 expression with or without block CDKN2A expression, a diagnosis of low-grade serous tumors is highly unlikely. In contrast, the positive predictive value is only 68%; hence, the test result alone cannot be used to diagnose low-grade serous tumors. Classical morphological features (uniform nuclei, micropapillary architecture, and low mitotic count) form the basis for a diagnosis of low-grade serous tumors with the appropriatewild-type TP53 and CDKN2A patchy staining pattern, serving as confirmation.
The data also show that TP53 confers most of the discriminatory information, most likely because of the almost universal prevalence of TP53 mutations in high-grade serous carcinomas6, 7 and absence in low-grade serous tumors. The specificity for low-grade serous tumors increases only from 89 to 93% by adding CDKN2A. Therefore, it points to the importance of correctly titrated TP53 immunohistochemistry and correct interpretation of the staining by pathologists. Defining TP53 positivity by an arbitrary cutoff such as 10% makes no sense considering the biological (ie, mutational) correlate for the aberrant or ‘all or nothing’ staining pattern.23, 26, 28 It has been previously shown that diffuse strong TP53 overexpression in at least 50–60% of the tumor cells is indicative of underlying missense mutation, resulting in accumulation of the protein. Complete absence of staining indicates null mutation, in which a deleterious mutation leads to mRNA, which is degraded by nonsense-mediated decay.26 Further studies to confirm these associations are warranted. Interpretation of CDKN2A staining is also critical. Similar to human papillomavirus-induced neoplasms of the uterine cervix, a high cutoff, likely close to 100% of tumor cells, should be considered as positive. We recently reported that CDKN2A is subject to a certain degree of intratumoral heterogeneity in high-grade serous carcinomas, which could lead to different results in 15–20% of cases.24 This is not the case for TP53. Therefore, one could consider CDKN2A as a safeguard in this panel, which is most useful in the setting of a wild-type TP53-staining pattern (clusters 2 and 5), whereby, it helps prevent the misdiagnosis of low-grade serous tumors in rare cases of high-grade serous carcinomas with a wild-type TP53 pattern (cluster 2), and supports a diagnosis of low-grade serous tumors in cluster 5. A three-tier approach to the interpretation of CDKN2A immunohistochemistry, which includes a separate category for absent CDKN2A staining, similar to TP53, is not indicated, as our data show that similar numbers of low-grade serous tumors and high-grade serous carcinomas are completely negative for CDKN2A (about 5%), similar to previous reports.7, 29
Additional molecular markers for low-grade serous tumors have limited diagnostic value. Next-generation sequencing of seven low-grade serous carcinomas did not reveal other recurrent mutations beyond KRAS and BRAF in a recent series, suggesting that low-grade serous carcinomas is a mutationally sparse disease30 Our result for KRAS mutation frequency is similar to the approximate 20% that had been previously reported.4 Although some reports have shown BRAF mutations in about 30% of cases,4, 30 our inability to detect BRAF V600E mutation in a single low-grade serous carcinomas is in line with a previous report in which Wong et al.31 assessed 43 advanced stage low-grade serous carcinomas. KRAS mutations were detected in 19% of cases, whereas BRAF mutations were found in only 2% of cases, which is very similar to our results. This suggests that serous borderline tumors with KRAS mutations are more likely to progress to low-grade serous carcinomas than serous borderline tumors with a BRAF V600E mutation. Similar results have been demonstrated in animal studies, where KRAS-mutant mice were much more likely to develop invasive lung adenocarcinoma than those with BRAF V600E mutations, which almost never spontaneously progressed to adenocarcinoma.32, 33 If the clinical trial, GOG239, confirms a differential mutation frequency between KRAS and BRAF in low-grade serous carcinomas, mutation profiling may prove to be a useful adjunct to morphology in the distinction between serous borderline tumors and low-grade serous carcinomas in small specimens (peritoneal cytology, small biopsies), which is critical for future neoadjuvant chemotherapy treatment decisions or clinical trial enrollment.
What is the current and future clinical relevance of making a distinction of high-grade serous carcinomas versus low-grade serous tumors, ie, vs low-grade serous carcinomas or serous borderline tumors? A recent clinical trial demonstrated that initial neoadjuvant chemotherapy followed by interval debulking was equivalent in outcome compared with the traditional upfront debulking surgery followed by adjuvant chemotherapy in advanced ovarian carcinoma, but with significantly less perioperative morbidity, mortality, and costs associated with neoadjuvant chemotherapy.34, 35 Neoadjuvant chemotherapy is, therefore, quickly becoming the favoured approach for advanced stage ovarian cancer. However, there are histological types of ovarian carcinoma that do not respond to chemotherapy as compared with the typical high-grade serous carcinomas. Low-grade serous carcinomas have been shown to be one of the histological types that shows a poor response to neoadjuvant chemotherapy.13, 15, 36 Five out of six low-grade serous carcinomas in the current Tom Baker Cancer Centre cohort were classified as chemotherapy resistant. Although low-grade serous carcinomas are relatively uncommon, it comprise the second most-common histotype in the setting of advanced stage ovarian carcinoma after high-grade serous carcinomas.37 A robust tissue-based diagnosis of low-grade serous carcinomas prior to any treatment could influence the decision making of gynecological oncologists in favor of upfront surgery to spare these women from neoadjuvant chemotherapy. However, current clinical practice does not rely on the acquisition of solid tumor tissue for the commencement of neoadjuvant chemotherapy. The results of the current study are based on tissue microarray cores (two cores each 0.6 mm), which is similar to the amount of tissue derived by a core biopsy, making these results fully referable if the same fixation and processing protocols are used. However, in most current clinical settings, diagnosis is based on more easily obtainable cytology. There is no reason to believe that the results obtained from collections of ascites fluid may differ from the current study. Our results on 30 cases support that the marker panel can be reliably interpreted on cell blocks using the same fixatives and immunohistochemical protocols. We would propose that the two marker panel TP53 and CDKN2A, in conjunction with PAX8 and WT1, in order to establish müllerian and serous cell lineage upfront, is highly effective in distinguishing between low-grade serous tumors and high-grade serous carcinomas in small tissue samples.
Clinical trials specifically targeting the critical molecular alterations of low-grade serous carcinomas are ongoing or in the planning phase.15 If patients are going to benefit from molecular-targeted therapy as shown for other cancer types with similar molecular alterations,38, 39, 40 robust diagnosis using ancillary markers is critical.
References
Burks RT, Sherman ME, Kurman RJ . Micropapillary serous carcinoma of the ovary. A distinctive low-grade carcinoma related to serous borderline tumors. Am J Surg Pathol 1996;20:1319–1330.
Seidman JD, Cho KR, Ronnett BM et al. Surface epithelial tumors of the ovary In: Kurman RJ, Ellenson LH, Ronnett BM (eds). Blaustein's Pathology of the Female Genital Tract 6th edn Springer: New York, 2011, pp 705–726.
Singer G, Kurman RJ, Chang HW et al. Diverse tumorigenic pathways in ovarian serous carcinoma. Am J Pathol 2002;160:1223–1228.
Singer G, Oldt R, Cohen Y et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst 2003;95:484–486.
Köbel M, Huntsman D, Gilks CB . Critical molecular abnormalities in high-grade serous carcinoma of the ovary. Expert Rev Mol Med 2008;10:e22.
Ahmed AA, Etemadmoghadam D, Temple J et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 2010;221:49–56.
Bell D, Berchuck A, Birrer M et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–615.
Malpica A, Deavers MT, Lu K et al. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol 2004;28:496–504.
Malpica A, Deavers MT, Tornos C et al. Interobserver and intraobserver variability of a two-tier system for grading ovarian serous carcinoma. Am J Surg Pathol 2007;31:1168–1174.
Vang R, Shih IeM, Kurman RJ . Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol 2009;16:267–282.
Gershenson DM, Sun CC, Lu KH et al. Clinical behavior of stage II-IV low-grade serous carcinoma of the ovary. Obstet Gynecol 2006;108:361–368.
Schmeler KM, Sun CC, Bodurka DC et al. Neoadjuvant chemotherapy for low-grade serous carcinoma of the ovary or peritoneum. Gynecol Oncol 2008;108:510–514.
Gershenson DM, Sun CC, Bodurka D et al. Recurrent low-grade serous ovarian carcinoma is relatively chemoresistant. Gynecol Oncol 2009;114:48–52.
Schmeler KM, Sun CC, Malpica A et al. Low-grade serous primary peritoneal carcinoma. Gynecol Oncol 2011;121:482–486.
Diaz-Padilla I, Malpica AL, Minig L et al. Ovarian low-grade serous carcinoma: a comprehensive update. Gynecol Oncol 2012;126:279–285.
Köbel M, Kalloger SE, Baker PM et al. Diagnosis of Ovarian Carcinoma Cell Type is Highly Reproducible: A Transcanadian Study. Am J Surg Pathol 2010;34:984–993.
Kalloger SE, Köbel M, Leung S et al. Calculator for ovarian carcinoma subtype prediction. Mod Pathol 2011;24:512–521.
Kommoss S, Pfisterer J, Reuss A et al. Specialized pathology review in patients with ovarian cancer: results from a prospective study. Modern Pathol 2012;25 (Suppl):281A.
Freedman OC, Dodge J, Shaw P et al. Diagnosis of epithelial ovarian carcinoma prior to neoadjuvant chemotherapy. Gynecol Oncol 2010;119:22–25.
Singer G, Stöhr R, Cope L et al. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol 2005;29:218–224.
O'Neill CJ, McBride HA, Connolly LE et al. High-grade ovarian serous carcinoma exhibits significantly higher p16 expression than low-grade serous carcinoma and serous borderline tumour. Histopathology 2007;50:773–779.
Köbel M, Kalloger SE, Boyd N et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 2008;5:e232.
Köbel M, Reuss A, Bois AD et al. The biological and clinical value of p53 expression in pelvic high-grade serous carcinomas. J Pathol 2010;222:191–198.
Köbel M, Turbin D, Kalloger SE et al. Biomarker Expression in Pelvic High-grade Serous Carcinoma: Comparison of Ovarian and Omental Sites. Int J Gynecol Pathol 2011;30:366–371.
Bromley AB, Altman AD, Chu P et al. Architectural Patterns of Ovarian/Pelvic High-grade Serous Carcinoma. Int J Gynecol Pathol 2012;31:397–404.
Yemelyanova A, Vang R, Kshirsagar M et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol 2011;24:1248–1253.
Capper D, Berghoff AS, Magerle M et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol 2012;123:223–233.
McCluggage WG, Soslow RA, Gilks CB . Patterns of p53 immunoreactivity in endometrial carcinomas: ‘all or nothing' staining is of importance. Histopathology 2011;59:786–788.
Schlosshauer PW, Deligdisch L, Penault-Llorca F et al. Loss of p16INK4A expression in low-grade ovarian serous carcinomas. Int J Gynecol Pathol 2011;30:22–29.
Jones S, Wang TL, Kurman RJ et al. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol 2012;226:413–420.
Wong KK, Tsang YT, Deavers MT et al. BRAF mutation is rare in advanced-stage low-grade ovarian serous carcinomas. Am J Pathol 2010;177:1611–1617.
Dankort D, Filenova E, Collado M et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 2007;21:379–384.
Trejo CL, Juan J, Vicent S et al. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res 2012;72:3048–3059.
Vergote I, Tropé CG, Amant F et al. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N Engl J Med 2010;363:943–953.
Hoskins PJ . Which is the better surgical strategy for newly diagnosed epithelial ovarian cancer: primary or interval debulking? Curr Opin Oncol 2011;23:501–506.
Schmeler KM, Gershenson DM . Low-grade serous ovarian cancer: a unique disease. Curr Oncol Rep 2008;10:519–523.
Köbel M, Kalloger SE, Huntsman DG et al. Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int J Gynecol Pathol 2010;29:203–211.
Gershenson DM, Sun CC, Iyer RB et al. Hormonal therapy for recurrent low-grade serous carcinoma of the ovary or peritoneum. Gynecol Oncol 2012;125:661–666.
Chapman PB, Hauschild A, Robert C et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–2516.
Prahallad A, Sun C, Huang S et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–103.
Acknowledgements
We thank Shuhong Liu (Anatomical pathology research laboratory at Calgary Laboratory Services) for tissue microarray construction and immunohistochemical stains. The study was funded by an internal Calgary Laboratory Research Competition (RS11-508).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Altman, A., Nelson, G., Ghatage, P. et al. The diagnostic utility of TP53 and CDKN2A to distinguish ovarian high-grade serous carcinoma from low-grade serous ovarian tumors. Mod Pathol 26, 1255–1263 (2013). https://doi.org/10.1038/modpathol.2013.55
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.55
Keywords
This article is cited by
-
A comprehensive immunohistochemical analysis of 26 markers in 250 cases of serous ovarian tumors
Diagnostic Pathology (2023)
-
Markers of MEK inhibitor resistance in low-grade serous ovarian cancer: EGFR is a potential therapeutic target
Cancer Cell International (2019)
-
High-grade serous ovarian carcinoma with mucinous differentiation: report of a rare and unique case suggesting transition from the “SET” feature of high-grade serous carcinoma to the “STEM” feature
Diagnostic Pathology (2019)