Abstract
Recent reports have indicated human cytomegalovirus (HCMV) to be associated with human glioblastoma carcinogenesis. In established examples of viral carcinogenesis, viral DNA and one or more of its products have been detected in most tumor cells of biopsies in the majority of cases. To test whether HCMV is associated with human glioblastoma based on this criterion, we measured the number of viral DNA molecules per cell in both frozen and paraffin-embedded tumor biopsies from 58 patients using real-time quantitative PCR (QPCR). Immunohistochemical and fluorescence in situ hybridization (FISH) to detect HCMV proteins and genome was performed in 10 cases using formalin-fixed paraffin-embedded glioblastoma tissues. Southern blotting using DNA extracted from four glioblastoma cell lines together with immunoblotting using the four cell lines and five glioblastoma tissue samples were also performed. We further confirmed the immunoblot bands using liquid chromatography-tandem mass spectrometry assay. As a result, HCMV DNA was not detected in the tumor cells from any of the glioblastoma cases by QPCR detecting two different HCMV genes, in clear contrast to samples from patients with HCMV infection. Southern blotting and immunoblotting of cell lines and FISH using paraffin sections were all negative. However, immunoblotting and immunohistochemistry using tissue samples were partly positive, but HCMV proteins were not detected by proteomic analysis, suggesting false positivity of the analyses. As our QPCR analysis could detect 10 copies of HCMV DNA mixed with DNA extracted from 104 HCMV-negative cells, we conclude that HCMV is not persistent, at least in the tumor cells, of developed human glioblastoma.
Similar content being viewed by others
Main
Human cytomegalovirus (HCMV) is a ubiquitous beta-herpes virus for which primary infection is generally asymptomatic, leading to latent infection during childhood. The virus is sometimes reactivated under immunosuppressive conditions, causing pneumonitis, hepatitis, retinitis and gastrointestinal diseases. Recently, HCMV was reported to be associated with most cases of human glioblastoma, in which viral DNA, RNA and proteins were all positive in the majority of the tumor cells.1 HCMV has also been detected in anaplastic gliomas and low-grade glioma.2 One group reported positivity in 10% of tumors using immunohistochemistry and fluorescence in situ hybridization (FISH).3 Slinger et al4 reported that tumor specimens from glioblastoma patients demonstrated colocalization of the HCMV encoded G protein-coupled receptor, US28, and phosphorylated STAT3 in the vascular niche. The data suggest that HCMV proteins may have a role in genesis of glioblastoma and immunotherapy studies for glioblastoma targeting viral proteins have been attempted in several laboratories. However, Lau et al5 did not detect any association between HCMV and glioblastomas by conventional PCR and other methods, and another group reported no involvement6 after analyzing a large number of samples. The role of methodology may be important and Cobbs et al1 observed that almost all primary tumor specimens from patients with glioblastoma multiforme expressed HCMV immediate early (IE) proteins, whereas adjacent normal cells did not. It is known that the HCMV 72-kDa IE protein (IE72) can enhance cell proliferation, thereby promoting cellular transformation or tumor progression.7 UL97 protein kinase phosphorylates and inactivates Rb, leading to cellular proliferation,8 and HCMV infection also is reported to activate human telomerase, preventing replicative senescence.9 In addition, the US28 protein promoted tumorigenesis when US28-expressing cells were injected into nude mice. Nevertheless, a causal relationship between HCMV infection and cancer has not been established and whether HCMV actually contributes to human glioblastoma oncogenesis in vivo is still unclear.10
Human papilloma virus and Epstein–Barr virus are well-established examples of viral oncogenesis with human malignancies (reviewed by Psyrri and DiMaio11 and Shah and Young12). In such cases, viral DNA and proteins are detected in the majority of the tumor cells in a large proportion of cases. If HCMV is the most important risk factor for the genesis of glioblastomas, it might similarly be expected to be detected in tumor cells, that is, the majority of tumor cells within a biopsy each have one or more copies of the HCMV genome.
Real-time quantitative PCR (QPCR) using HCMV-specific primers and appropriate probes is well established for testing viral load of HCMV in clinical samples,13 with high specificity and sensitivity.14 Using this method, at least one copy of HCMV genome per 1000 cells can be detected.15
In this study, we quantitatively measured the number of viral DNA molecules per cell in both frozen and paraffin-embedded tumor biopsies from 58 glioblastoma patients using QPCR. As a result, no positive signals could be detected, and we attempted to perform proteome analysis to identify which proteins contributed to the signals frequently detected in the glioblastoma tumor cells by immunoblotting and immunohistochemistry.
Materials and methods
Patients
Frozen glioblastoma tissues (40 samples) were obtained from 25 patients of Nagoya University Hospital and 15 of Nagoya Central Hospital with written informed consent. In addition, 19 formalin-fixed, paraffin-embedded glioblastoma samples were obtained from 10 patients of Nagoya University Hospital and 9 of Toyota Kosei Hospital. The patients were 36 men and 22 women, aged 12–84 (mean 55.7, median 61). For positive controls, fresh blood or urine was obtained from two pediatric patients with HCMV infection. Furthermore, formalin-fixed, paraffin-embedded samples were obtained from two adult patients with HCMV reactivation. Cell lysates from fresh frozen tissues from patients #1, 3, 4, 8, 18 in Table 1 were applied for immunoblotting. Paraffin sections from patients #41–50 and #P3 in Table 1 were applied for immunohistochemistry and FISH. The experimental designs in this study were reviewed and approved by the Committee for Bioethics of Nagoya University Graduate School of Medicine (#671 and #830) and Nagoya Central Hospital.
Cell Lines
Human peritoneal mesothelial cells (HPMCs) were established and cultured as previously described.16 Human foreskin fibroblasts were cultured and infected by HCMV as previously described.17 The glioblastoma cell lines, T98, U87, SKKG7 and G251 were maintained in Eagle’s minimal essential medium supplemented with 10% fetal calf serum (Sigma, St Louis, MO, USA), penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37 °C in 5% CO2. These four glioblastoma cell lines were applied for Southern blotting and immunoblotting analyses.
DNA Extraction
DNA was extracted from 10 μm paraffin-embedded sections using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA). From frozen tissue samples or cells in culture, genomic DNA was directly extracted using a QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. Towne18 HCMV bacterial artificial chromosome (BAC) DNA including the whole genome of HCMV was kindly donated by F Liu (University of California, Berkeley, CA, USA). Plasmid containing IE gene of HCMV was constructed as previously described. Plasmid containing glycoprotein B (gB) gene was constructed by first amplifying the gB gene with the primers described,19 and cloned into the pGEM-T Easy TA cloning kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. BAC or plasmid DNA was extracted using a Nucleobond PC 20 Plasmid DNA Purification Kit (Macherery-Nagel, Düren, Germany) or a plasmid midi prep kit (Qiagen), respectively, according to the manufacturer’s instructions.
Real-Time QPCR
Fifty nanograms of isolated genomic DNA were applied per each QPCR reaction. Sample DNA was amplified 50 cycles with an annealing temperature of 55 °C using a Taqman Universal PCR Kit (ABI) and the 7300 real-time PCR system (ABI) following the manufacturer’s instructions. PCR products were normalized to human beta-actin using the comparative Ct method. Water was used to test for DNA contamination in the reagents. The primers and probes for gB and major IE gene.17, 19
Immunoblotting and Immunohistochemical Analyses
Immunoblotting and immunohistochemistry were performed as previously described.20, 21 Purified human serum albumin (HSA) was obtained from Wako Pure Chemical Industries (#013-10501). Primary antibodies used were IE1/2 (NEA-9221, Perkin Elmer), pp28 (CA004, EastCoast Bio), pp65 (023-A, Virogen) and UL44 (CA006, EastCoast Bio).
Liquid Chromatography-Tandem Mass Spectrometry (LC/MS/MS) Analysis
For LC/MS/MS analysis, bands were visualized using 4-chloro-1-naphtol, excised and then on-membrane digestion was performed mostly similarly to a previous report.22 LC/MS/MS analysis was performed as previously described.23
Fluorescence In Situ Hybridization
HCMV BAC DNA was labeled with SpectrumOrange using a Nick Translation Kit (Abbott Molecular, Abbott Park, IL, USA) according to the manufacturer’s recommendation and FISH analysis for HCMV genome detection was performed as described previously.24
Southern Blot Analysis
A HCMV probe was generated by labeling HCMV BAC DNA with [32P]dCTP (Amersham Pharmacia Biotech, Piscataway, NJ, USA) using a Nick Translation Kit and Southern blot analysis was performed as described previously.17
Results
QPCR and Southern Blot Analysis
Clinical data of the patients and results for detection of HCMV DNA are summarized in Table 1. DNA from samples #1–40 and 41–59 were extracted from fresh-frozen, and formalin-fixed paraffin-embedded tissues, respectively. Samples #6 and 49 were obtained from a single patient. The primers and probes were validated with DNA extracted from the plasmid containing IE and gB genes of HCMV, HCMV BAC DNA and HCMV BAC DNA admixed with cellular DNA. The minimum detection limits of plasmids encoding HCMV IE gene or gB gene, HCMV-BAC, and HCMV-BAC admixed with HCMV-negative cellular DNA isolated from HPMC cells were 2 copies, 1 copy, 1 copy and 10 copies mixed with 10 000 cells per reaction, respectively. Cell numbers for each sample were estimated by comparing the amounts of beta-actin PCR product with 1 × 104 HPMC cells. The estimated number of cells per reaction of the glioblastoma samples varied from 85 to 104 527 cells (median 17 073, mean 21 352; Table 1). The HCMV copy numbers for urine or blood from two patients with symptoms of HCMV hepatitis and systemic HCMV infection were 39 and 12 336 per reaction, respectively (samples P1 and P2) and 394 and 8826 per reaction, respectively, for formalin-fixed paraffin-embedded samples from two other patients with CMV proctitis or gastritis (samples P3 and P4). The minimum HCMV copy number was 0.002 per cell in these positive control samples. In clear contrast, HCMV DNA for either gB or IE genes was not detected in any of the 59 glioblastoma samples. To confirm that formalin-fixed DNA from HCMV-negative glioblastoma samples, we performed control experiments using 10 000, 1000, 100, 10, 5, 2 and 1 copy (copies) of HCMV BAC DNA admixed in 50 ng of HCMV-negative samples (#41). Positive results in triplicate were detected in samples containing >10 copies of HCMV DNA in both IE and gB primer and probes, but no repetitive positive result were obtained from samples containing five HCMV copies or less. Southern blot analysis was totally negative using DNA extracted from cell lines (Supplementary Figure S1).
Immunoblotting and LC/MS/MS
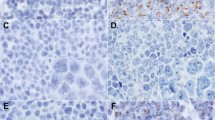
Immunoblotting of proteins extracted from five Japanese glioblastoma cases revealed that relatively clear, positive bands were obtained using IE1 and pp28 antibodies (Supplementary Figure S2A). Thus, we further performed LC/MS/MS analysis of the excised bands. From the band detected with the IE1 antibody, 69 kDa-sized HSA was specifically detected recurrently with high scores (>200), and from the band detected with the pp28 antibody, peptides of isomers of myelin basic protein (MBP) were most frequently detected with reliable scores (Supplementary Table S2). Viral proteins were considered as negative because of the low frequency and score. Thus, we performed immunoblotting using purified HSA and demonstrated that IE1 antibody could bind not only to glioblastoma lysates but also to purified HSA (Supplementary Figure S2B). Furthermore, no positive results were obtained using glioblastoma cell lines (Supplementary Figure S2C). However, we obtained vague to rather intense positive nuclear staining of viral antigens by immunohistochemistry using antibodies against IE1 and pp28 of HCMV (Figure 1).
Immunohistochemical and fluorescent in situ hybridization analyses of glioblastoma. (a–e). Glioblastoma (#41 in Supplementary Table S1 and S2). (f–k). Reactive HCMV proctitis (#P3 in Table 1). H&E stain (a, f), immunostaining (b, g: IE1/2; c, h: pp28; d, i; pp65, e, j; UL44), FISH using a HCMV whole genome probe labeled orange (k). (a) Glioblastoma cells with pleomorphic nuclei surrounding a necrotic focus are seen. (b, c) Positive nucleic staining by IE1/2 and pp28 are seen in glioblastoma cells. (d, e) pp65 and UL44 were negative in this case (#41). (f) An HCMV-infected cell with perinuclear viral inclusion (owl’s eye) is seen. (g–k) The nucleus of HCMV-infected reactive cell is clearly stained by all HCMV antibodies (g–j) and FISH (k).
Immunohistochemical Analyses and FISH
Immunohistochemical results of 10 patients (#41–50 in Table 1) are summarized in Supplementary Table S1. Although some of the glioblastoma tumor cells showed rather strong nuclear staining using HCMV antibodies (Figures 1b and c), no staining was observed using others (Figures 1d and e), in clear contrast with the positive control sample from an HCMV proctitis patient in which intense nuclear staining was observed by all of the HCMV antibodies (Figures 1g–j). By FISH analysis, orange fluorescent signals were diffusely observed in the nuclei and perinuclear area of CMV-infected cells using paraffin sections of positive control samples (Figure 1k), however, no signals were detected in the 10 glioblastoma samples (#41–50 in Table 1 and Supplementary Table S1).
Discussion
Our QPCR assay demonstrated that the relative copy number of HCMV in all the frozen or formalin-fixed, paraffin-embedded tissues was <10 copies per 10 000 cells, that is, one HCMV genome in 1000 cells. The results are not in line with earlier reports that were interpreted to indicate that HCMV DNA was detectable in a fraction of human glioblastomas, despite the fact that the same region of the HCMV genome were amplified.25 Recently, another study using 12 primer pairs and semi-QPCR also revealed the significantly higher incidence of CMV-DNA in a proportion of glioblastoma samples compared with non-tumor tissues.26 One reason for this discrepancy may be because extremely small amount of DNA can be detected by traditional PCR techniques that were utilized such as nested-PCR utilized in the previous study1 in which 35–50 cycles of amplification followed by a second round of amplification, and HCMV DNA from cells other than the tumor cells were detected. Glioblastoma is often accompanied with reactive gliosis at the edge of the tumor tissue, and microglial cells that originate from bone marrow myeloid cells, which may be persistently infected by HCMV,27 could have important roles in the development of gliosis.28 Thus, we consider that these positive PCR results different from ours may be because of relatively smaller content of reactive glial cells in our samples. Another important reason may be because false positivity due to cross-contamination of the samples or incorporation of pre-existing HCMV DNA in the laboratory can easily occur using these PCR-based methods. In our study, negative control wells with no templates were always included in each PCR reaction plate to avoid over-interpretation of false-positive results.
Southern blotting and immunoblotting using DNA and protein extracted from glioblastoma cell lines both revealed negative results. Therefore, we consider that HCMV is negative in the neoplastic cells of developed glioblastoma. However, immunoblotting and immunohistochemistry using tissue samples both showed some positive signals, and nuclear staining was detected by immunohistochemistry not only in the tumor cells but also in the reactive glial cells (Supplementary Figure S2 and Supplementary Table S1). Recent studies suggest that HCMV infection may be important in inducing tumor-supportive monocytes to support the growth of the tumor cells,29 and also, DNA changes in single cells can influence other neighboring cells either through intracellular connections or paracrine mechanisms or exosomes,30 we speculated that even if HCMV does not exist in the tumor cells, HCMV infection may persist in the adjacent cells such as endothelial cells, monocyte-lineage cells or glial cells of reactive gliosis, and HCMV proteins are secreted into the tumor cells. However, we could not detect positive signals by FISH analysis within the tumor tissue (Supplementary Table S1). Our FISH analysis is highly sensitive because we probed BAC DNA containing full-length HCMV genome, and in our previous study, we could readily detect two copies of the human genome in a cell using the same method.24 Earlier studies reporting positive signals of HCMV DNA and RNA in the glioblastoma cells by in situ hybridization utilized antibody-mediated enhancement of the primary signals,1, 2 which may have resulted in augmentation of nonspecific bindings of the probes, leading to misinterpretation of the signals. Furthermore, our LC/MS/MS analysis revealed that proteins extracted from the positive bands detected in immunoblotting using IE1 and pp28 antibodies were non-viral human proteins such as HSA and MBP, suggesting previously unknown cross-reactivity of these antibodies and also the unreliability of not only positive immunoblot bands but also immunohistochemical staining. According to a most recent report performing RNA-Seq analysis using a next-generation sequencer on a large number of human tumor tissues, no viral RNA from DNA viruses such as HCMV was present in either low-grade or high-grade glial tumors.31 Taking into account, although there still is a possibility that HCMV-infected cells are present in the brain tissue at certain proportion, it is unlikely that the majority of the tumor cells are persistently infected, and we could not obtain direct evidences to support the hypothesis that exosome secretion of viral RNA and proteins from non-neoplastic cells are the reasons for positive immunoblotting or immunostaining results.
In conclusion, in this study, we analyzed a relatively large number of glioblastoma samples by QPCR, which revealed that all the cases were negative for HCMV viral DNA. Furthermore, we could not detect any viral proteins extracted from positive immunoblot bands using IE1 or pp28 antibody. Thus, we conclude that HCMV genome or proteins are not detectable in human glioblastoma cells.
References
Cobbs CS, Harkins L, Samanta M et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res 2002;62:3347–3350.
Scheurer ME, Bondy ML, Aldape KD et al. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol 2008;116:79–86.
Sabatier J, Uro-Coste E, Pommepuy I et al. Detection of human cytomegalovirus genome and gene products in central nervous system tumours. Brit J Cancer 2005;92:747–750.
Slinger E, Maussang D, Schreiber A et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal 2010;3ra58.
Lau SK, Chen YY, Chen WG et al. Lack of association of cytomegalovirus with human brain tumors. Mod Pathol 2005;18:838–843.
Poltermann S, Schlehofer B, Steindorf K et al. Lack of association of herpesviruses with brain tumors. J Neurovirol 2006;12:90–99.
Yonemitsu Y, Kaneda Y, Komori K et al. The immediate early gene of human cytomegalovirus stimulates vascular smooth muscle cell proliferation in vitro and in vivo. Biochem Biophys Res Comm 1997;231:447–451.
Hume AJ, Finkel JS, Kamil JP et al. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 2008;320:797–799.
Straat K, Liu C, Rahbar A et al. Activation of telomerase by human cytomegalovirus. J Natl Cancer Inst 2009;101:488–497.
Miller G . Brain cancer a viral link to glioblastoma? Science 2009;323:30–31.
Psyrri A, DiMaio D . Human papillomavirus in cervical and head-and-neck cancer. Nat Clin Pract Oncol 2008;5:24–31.
Shah KM, Young LS . Epstein-Barr virus and carcinogenesis: beyond Burkitt's lymphoma. Clin Microbiol Inf 2009;15:982–988.
Ito Y, Shibata-Watanabe Y, Kawada J et al. Cytomegalovirus and Epstein-Barr virus coinfection in three toddlers with prolonged illnesses. J Med Virol 2009;81:1399–1402.
Maeda N, Yamashita Y, Kimura H et al. Quantitative analysis of herpesvirus load in the lymph nodes of patients with histiocytic necrotizing lymphadenitis using a real-time PCR assay. Diagn Mol Pathol 2006;15:49–55.
Tanaka N, Kimura H, Iida K et al. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J Med Virol 2000;60:455–462.
Nagai H, Okazaki Y, Chew SH et al. Diameter and rigidity of multiwalled carbon nanotubes are critical factors in mesothelial injury and carcinogenesis. Proc Natl Acad Sci USA 2011;108:E1330–E1338.
Isomura H, Stinski MF . The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J Virol 2003;77:3602–3614.
Dunn W, Chou C, Li H et al. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci USA 2003;100:14223–14228.
Wada K, Kubota N, Ito Y et al. Simultaneous quantification of Epstein-Barr virus, cytomegalovirus, and human herpesvirus 6 DNA in samples from transplant recipients by multiplex real-time PCR assay. J Clin Microbiol 2007;45:1426–1432.
Yamashita Y, Kajiura D, Tang L et al. XCR1 expression and biased VH gene usage are distinct features of diffuse large B-cell lymphoma initially manifesting in the bone marrow. Am J Clin Pathol Apr 2011;135:556–564.
Yamashita Y, Tsurumi T, Mori N et al. Immortalization of Epstein-Barr virus-negative human B lymphocytes with minimal chromosomal instability. Pathol Int 2006;56:659–667.
Luque-Garcia JL, Zhou G, Spellman DS et al. Analysis of electroblotted proteins by mass spectrometry: protein identification after western blotting. Mol Cell Proteomics 2008;7:308–314.
Kobayashi H, Yamashita Y, Iwase A et al. The ferroimmunomodulatory role of ectopic endometriotic stromal cells in ovarian endometriosis. Fertil Steril 2012;98:415–422 e1-12.
Yamashita Y, Akatsuka S, Shinjo K et al. Met is the most frequently amplified gene in endometriosis-associated ovarian clear cell adenocarcinoma and correlates with worsened prognosis. PLoS One 2013;8:e57724.
Mitchell DA, Xie WH, Schmittling R et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro-Oncol 2008;10:10–18.
Ranganathan P, Clark PA, Kuo JS et al. Significant association of multiple human cytomegalovirus genomic loci with glioblastoma multiforme samples. J Virol 2012;86:854–864.
Khaiboullina SF, Maciejewski JP, Crapnell K et al. Human cytomegalovirus persists in myeloid progenitors and is passed to the myeloid progeny in a latent form. Brit J Haematol 2004;126:410–417.
Zhang D, Hu X, Qian L et al. Astrogliosis in CNS pathologies: is there a role for microglia? Mol Neurobiol 2010;41:232–241.
Dziurzynski K, Wei J, Qiao W et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Cancer Res 2011;17:4642–4649.
Meckes DG Jr., Shair KH, Marquitz AR et al. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA 2010;107:20370–20375.
Khoury JD, Tannir NM, Williams MD et al. The landscape of DNA virus associations across human malignant cancers using RNA-Seq: an analysis of 3775 cases. J Virol 2013;87:8916–8926.
Acknowledgements
We appreciate Ms Y Tanaka, Ms F Ando, Mr A Kamiya and Mr N Misawa for expert technical support. We are also grateful to Nagoya Central Hospital and Toyota Kosei Hospital for providing glioblastoma samples and clinical data. This work was supported by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology of Japan (no. 20390137, 21022055 to T Tsurumi), and partly by the Uehara Memorial Research Fund (to T Tsurumi).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Yamashita, Y., Ito, Y., Isomura, H. et al. Lack of presence of the human cytomegalovirus in human glioblastoma. Mod Pathol 27, 922–929 (2014). https://doi.org/10.1038/modpathol.2013.219
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.219
Keywords
This article is cited by
-
Cytomegalovirus DNA in non-glioblastoma multiforme brain tumors of infants
Child's Nervous System (2021)
-
Viral infection and glioma: a meta-analysis of prognosis
BMC Cancer (2020)
-
Human cytomegalovirus DNA and immediate early protein 1/2 are highly associated with glioma and prognosis
Protein & Cell (2020)
-
Cytomegalovirus and cancer-related mortality in the national health and nutritional examination survey
Cancer Causes & Control (2020)
-
Is the presence of HCMV components in CNS tumors a glioma-specific phenomenon?
Virology Journal (2019)