Abstract
Alterations in the retinoblastoma pathway are frequent in ovarian/tubal high-grade serous cancers, but the mechanism of deregulation and the impact on patient outcome are poorly understood. A cohort of 334 high-grade serous carcinomas was studied by immunohistochemical analysis of RB1, p16, cyclin D1, cyclin E1, and Ki67. Additional detailed analyses including RB1 allelic deletion (n=42), mutation (n=75), methylation (n=31), and SNP array analyses (n=75) were performed on cases with clinical parameters, including age, debulking status, treatment, and clinical outcome. p16/RB1 expression results yielded three distinct clinically relevant subgroups upon multivariable analysis controlling for stage, debulking status, and treatment types: p16 homogeneous/RB1+ with the shortest progression-free survival (median 15 months (95% CI: 13–18); P=0.016) compared with the p16 heterogeneous/RB1+ subgroup (median 22 months (95% CI: 16–32)) and the p16 homogeneous/RB1− subgroup (median 20 months (95% CI: 15–24)). Patients in the p16 homo/RB1− subgroup showed a significant increase in overall survival (>60 months; P=0.013), which suggests an increase in sensitivity to cytotoxic agents. Analyses of Rb pathway mechanistic differences among these groups revealed frequent RB1 genomic alterations such as RB1 allelic loss and/or large spanning deletions (83%) in the p16 homo/RB1− subgroups, also indicating that RB1 deletions are frequent in high-grade serous carcinoma. CCNE1 gene gains/amplifications were frequent in the p16 homogeneous/RB1+ subgroup (68%) and cyclin D1 protein overexpression was predominantly characteristic of the p16 heterogeneous/RB1+ subgroup. These subcategories occur early in tumor progression and are seen with similar frequency in the cancer precursor lesion, serous tubal intra-epithelial carcinoma. Overall, this study uniquely identifies multiple non-synonymous mechanisms of retinoblastoma pathway deregulation that correlate with significantly different clinical outcomes. Furthermore, deregulations identified in precursor lesions suggest a key role of this pathway in serous tumor development. Recognition of these categories may identify patients with increased sensitivity to chemotherapy and new opportunities for novel therapeutics.
Similar content being viewed by others
Main
High-grade serous cancer is the most common type of epithelial ovarian cancer, accounting for about 70% of all cases, and for the majority of epithelial ovarian cancer deaths.1, 2, 3, 4 Despite an initial high response rate to first-line adjuvant platinum-based chemotherapy and improvements in surgical and medical treatment options, the prognosis remains poor for most high-grade serous carcinoma patients.3, 4 Key challenges in improving overall survival include limited understanding of the early events in serous carcinogenesis, a lack of cancer screening strategies leading to over 90% diagnosis at an advanced stage of disease and ineffective treatment options for recurrent, platinum-resistant disease.2, 3, 4 Recent advances in molecular pathogenesis by genomic analyses demonstrate that high-grade serous carcinoma has a strikingly high prevalence of somatic copy number alterations, with few recurrent mutations.3, 4, 5 The spectrum of mutations is distinct from other types of epithelial ovarian cancer, reflecting the differing etiology, pathogenesis, and cell of origin in the different histotypes. In addition, deregulation of various pathways, including retinoblastoma, PI3K/Ras, homologous recombination, and Notch signaling pathways, may be used to further stratify high-grade serous carcinoma into molecular subgroups with potential relevance for use of targeted therapies.5, 6
The retinoblastoma pathway is an important regulator of proliferation at the G1/S checkpoint, and disruption of at least one arm of the retinoblastoma pathway is frequent and required in tumor formation/progression.7, 8, 9 Differential genetic and epigenetic mechanisms of retinoblastoma pathway disruption may lead to non-synonymous consequences dependent on tissue and tumor type.7, 8, 9, 10 Deregulation of key pathway members, such as cyclin D1 and cyclin E1 in breast carcinoma and RB1 and CDKN2A in lung carcinoma, predicts for clinical outcome and allows the differential alterations to be exploited when considering the efficacy of common and new therapeutic targets.7, 8, 9, 10
Aberrant CDKN2A elevation, identified by intense homogeneous expression, is seen in many cancers with loss of RB1 function, which has thus far been demonstrated to enable bypass of CDKN2A-mediated cell cycle arrest.11, 12 For example, p16 overexpression in cervical cancer is a surrogate of human papillomavirus infection (oncoprotein E7 targets RB1).13, 14 Similarly, elevated p16 occurs in 70% of high-grade serous carcinoma, but the RB1 mutation rate is low (4–6%) and only 6% of cases have protein loss.5, 15 Furthermore, the correlation between RB1 and p16 protein expression in high-grade serous carcinoma has not yet been explored in depth and associations between different retinoblastoma pathway alterations and clinical outcome/treatment response remain poorly investigated. In this study, we identify non-synonymous retinoblastoma pathway deregulation modalities that stratify patients with different responses to first-line platinum-based chemotherapy.
Materials and methods
Tissue Samples
The study protocol for collection of tissue and clinical information for all patients was approved by the UHN Research Ethics Board. All patients provided written informed consent authorizing collection and use of tissue for research purposes. Snap-frozen (n=75) and formalin-fixed paraffin-embedded (n=334) tissues were retrospectively selected from the University Health Network Biobank (1996–2008). Key inclusion criteria were high-grade serous tumors from patients naive to chemotherapy. Clinical annotation inclusive of age at diagnosis, stage, surgical debulking status, first-line chemotherapy, progression-free and overall survival was available for 210 cases (Supplementary Table 1). Progression-free survival was defined as the time elapsed between diagnosis and progression, based on the first confirmed sign of disease recurrence. Overall survival was defined as the time elapsed between diagnosis and death by disease or date of last follow-up.
Immunohistochemistry
Protein expression for p16 (CINtec® Histology V-Kit, Roche mtm Laboratories) and RB1 (NCL-RB1, Novacastra) was assessed by immunohistochemistry using standard techniques on tissue microarrays composed of 334 high-grade serous carcinoma formalin-fixed paraffin-embedded tissues.
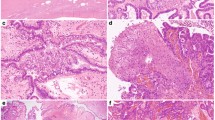
p16 expression was divided into three categories: 0=negative, 1= heterogeneous staining (patchy staining in three tumor fields), 3=diffuse homogeneous staining (100% staining in three tumor fields; Figure 1a). RB1 protein expression was divided into a positive and negative stain (defined by complete absence of signal; Figure 1b). Protein expression for cyclin E1, cyclin D1, and Ki67 was also determined (Supplementary Table 2).
Kaplan–Meier curves of the differences identified in progression-free survival and overall survival based on p16/RB1 immunohistochemical stratification. Representative images of immunohistochemical scoring for (a) p16—negative, heterogeneous, homogeneous; (b) RB1—negative and positive ( × 20 magnification). Patient subgroups with differential p16 expression patterns showed significant differences in progression-free survival and overall survival (c) and (d). Patient subgroups with differential p16/RB1 expression patterns also showed significant differences in progression-free survival and overall survival (e) and (f).
RB1 Allelic Loss, Mutation, Methylation, and Sequencing Analyses
These analyses were performed in collaboration with Impact Genetics, a clinical testing laboratory that has optimized molecular assays to identify RB1 mutations. Patient genomic DNA was isolated from normal fallopian tube/ovarian and high-grade serous carcinoma tumor tissues using the DNeasy kit (Qiagen, Canada). Quantitative multiplex PCR was performed on 75 patient samples to detect deletions, duplications, or insertions, and exon copy number in exons 1–27 with flanking introns and the promoter region. Allele-specific PCR was performed to detect SNPs at 11 locations based on known recurrent nonsense RB1 mutations and RB1 promoter methylation status was determined on 31 tumors as previously described.16, 17, 18 For RB1 allelic analyses (n=42), three polymorphic markers were used: a variable number of tandem repeats in intron 20 (RB1.2) and two microsatellite markers D13S153 and D13S164 (Supplementary Figure 1D). A sample marker was called ‘informative’ when two alleles with a different number of variable number of tandem repeats were present in the normal DNA. RB1 allelic loss at a particular polymorphic region was determined based on the height differences of the cancer and normal control peaks using the allelic imbalance ratio (AIR)=(T1)*(N2)/(T2)*(N1), from Banks et al., where allelic loss=AIR>1.2, AIR<0.8.19
Statistical Analysis
Associations between the p16/RB1 subgroups and clinical outcome were examined using Kaplan–Meier survival analysis and log-rank test. Multivariable analyses for progression-free survival and overall survival were performed to control for debulking status, stage, and treatment using the Cox model. Associations between copy number/mutations/allelic loss alterations of Rb pathway members and the p16/RB1 subgroups were identified using Pearson χ2 analyses. To determine differential expression of cyclin D1, cyclin E1, and Ki67 across the identified RB1/p16 subgroups, ANOVA and Tukey’s honest Range significance tests were performed. SPSS 14.0 (SPSS, Chicago, IL, USA) and SAS 9.2 were used to perform these analyses.
Results
p16/RB1 Protein Expression Patterns Define Clinical Outcome in High-Grade Serous Carcinoma
Immunohistochemical analyses for p16 and RB1 with three fields assessed per tumor were performed on a cohort of 334 high-grade serous carcinoma cases with a median follow-up time of 41 months (95% CI: 38–45). p16 expression was characterized by three patterns: negative (6%), heterogeneous (31%), and homogeneous (63%; Table 1, Figure 1a). Patients with heterogeneous expression showed a significantly longer progression-free survival (median 22 months (95%CI:16–32)); P=0.028) and overall survival (median 66 months (95% CI:38–86)); P=0.021) (Table 1, Figure 1c and d). RB1 immunohistochemistry identified two subgroups, negative (33%) and positive (67%; Table 1, Figure 1b). There was no significant difference between these groups in age, progression-free survival (median 20 vs 18 months, respectively; P=0.339), or overall survival (median 44 vs 42 months, respectively, P=0.720; Table 1).
Next, we analyzed the effects on clinical outcome of combining p16 and RB1 protein expression patterns for all high-grade serous carcinoma cases, where six subgroups were analyzed, the most prevalent being: p16 homogeneous/ RB1 positive (p16 homo/RB1+) at 34%, p16 heterogeneous/RB1 positive (p16 hetero/RB1+) at 27% and p16 homogeneous/RB1 negative (p16 homo/RB1−) at 28% of all samples (Table 1). The median progression-free survival and overall survival for the poor outcome group (p16 homo/ RB1+) were 15 months (95% CI: 13–18) and 39 months (95% CI: 35–44; Table 1). Patients in the p16 homo/RB1+ subgroup showed a shorter progression-free survival compared with the p16 hetero/RB1+ and p16 homo/RB1− subgroup (median 22 vs 20 months, respectively, P=0.003; Table 1, Figure 1e). Likewise, the p16 homo/RB1+ subgroup showed a shorter overall survival compared with the p16 hetero/RB1+ and p16 homo/RB1− (median 66 vs 44 months, respectively, P=0.013; Table 1, Figure 1f). Following multivariable analyses, considering predictors of outcome, such as debulking status (optimal/sub-optimal), treatment (platinum vs platinum and taxane), and stage (low/high), significantly shorter progression-free survival (P=0.016) and overall survival (P=0.049) was found in the p16homo/RB1+ subgroup compared with the p16 hetero/RB1+ and the p16 homo/RB1− subgroups (Table 2).
RB1 Hemizygous Deletions Are Frequent in High-Grade Serous Carcinoma and Correlate with RB1 Functional Loss
Forty-two high-grade serous carcinoma tumors and their corresponding normal tissues were analyzed for RB1 allelic deletions at three polymorphic regions (D13S153, RB1.20, D13S164) located on chromosome 13q14.2; thirty-eight samples were informative for at least one marker (Supplementary Figure 1D). Twenty-nine out of thirty-eight (76%) samples had allelic loss at either D13S153 or RB1.20 and fifteen out of twenty-nine (52%) samples correlate with a loss of RB1 protein expression (Figure 2a). Furthermore, 21/29 (72%) of samples with RB1 allelic loss have p16 homogeneous expression (Figure 2a). These findings show that RB1 allelic loss is prevalent in high-grade serous carcinoma and that it correlates with p16 overexpression.
Deregulations at the RB1 gene (chromosome 13q14.2). (a) Correlations between RB1 allelic loss and the three clinically distinct p16/RB1 subgroups. (b) Correlations between RB1 copy number alterations and the three clinically distinct p16/RB1 subgroups. (c) Summary of all RB1 gene alterations illustrating a high frequency of whole gene or large spanning gene deletions and few point mutations.
The cohort of 75 high-grade serous carcinoma samples was utilized for a thorough investigation of the RB1 gene, by performing copy number, mutation, and hypermethylation analyses. RB1 hemizygous deletions were identified in 38% of samples, however, no homozygous deletions were detected by SNP analysis and RB1 copy number gains were identified in only 6% samples (Supplementary Figure 1A and B). Functional loss associated with RB1 hemizygous deletions, as 71% (20/28) of samples, showed downregulation of RB1 mRNA (P=0.0008) and a loss of protein expression (Supplementary Figure 1C and Supplementary Table 3).
Mutation analyses (quantitative multiplex-PCR and allele specific-PCR) revealed 40 different RB1 gene alterations across the cohort: 30 exonic deletions, 1 homozygous single-base mutation, and 9 exonic amplifications (Figure 2c). Mapping of the deletions and amplifications onto RB1 protein domains did not reveal any particular mutation hotspots, as most span the entire gene region or are multi-exonic deletions (Figure 2c). In the p16 homo/RB1− subgroup, 26/31 (83%) of samples resulted in functional loss of protein, including the homozygous mutation (R445X; Figure 2b and c). Strong synergy was found between SNP and mutation analyses in terms of RB1 deletions identified. Methylation analyses on all 31 p16 homo/RB1− group samples revealed no promoter hypermethylation (data not shown).
p16/RB1 Subgroups Show Characteristic Deregulations of the Retinoblastoma Pathway
Copy number alterations were found in all seven key retinoblastoma pathway members with correlative changes in mRNA expression in five members: RB1, CDKN2A, CCNE1, E2F3, and CDK4 (Supplementary Table 3 and Supplementary Figure 1B). CCNE1 overexpression is a striking feature of p16 homo/RB1+, with copy number gains and amplifications in 68% (13/19; P=0.005; Figure 3c) and showed significant protein overexpression compared with the other p16/RB1 subgroups (P<0.05; Figure 3a and b), suggesting that genomic gain/amplification of this oncogene has an important role in this subgroup. Another trend was also observed with E2F3, where all five amplifications and many of the copy number gains (10/29) where identified in p16 homo/RB1− subgroup samples (P=0.08).
Immunohistochemical quantification of cyclin E1, cyclin D1, and Ki67 protein expression and quantification of CCNE1 and CCND1 CNA across the p16/RB1 clinical subgroups. (a) Immunohistochemical representation of cyclin E1, cyclin D1, and Ki67 ( × 20 magnification). (b) Protein expression quantification of cyclin E1, cyclin D1, and Ki67; (c) copy number alterations (CNA) of CCNE1 and (d) CCND1 across the p16/RB1 subgroups (*P<0.05, **P<0.01).
Cyclin D1 immunohistochemistry on 334 samples revealed predominant protein overexpression in the p16 hetero/RB1+ subgroup (P<0.05; Figure 3a and b), although CCND1 copy number alterations did not correlate with CCND1 mRNA expression (Supplementary Table 3) or with any particular p16/RB1 subgroup (Figure 3d). In addition, cyclin D1 protein expression observed in the p16 hetero/RB1+ subgroup was significantly higher than in the normal fallopian tube epithelium (Supplementary Figure 3). These results suggest that cyclin D1 overexpression, does occur in high-grade serous carcinoma, is most frequent in the p16 hetero/RB1+ subgroup and most likely occurs as a result of post-translational modifications. Interestingly, the p16 hetero/RB1+ subgroup contained a large proportion of samples without any retinoblastoma pathway copy number alterations, indicating that patients in this cohort are largely retinoblastoma pathway neutral except for those with cyclin D1 overexpression.
We also analyzed Ki67 immunohistochemistry in the 334 high-grade serous carcinoma samples to estimate proliferative index. The p16 homo/RB1− subgroup exhibited the highest Ki67 expression compared with either p16 hetero/RB1+ (P<0.05) or p16 homo/RB1+ subgroups (P<0.05; Figure 3a and b). Overall, high-grade serous carcinoma by definition are considered to have a high mitotic rate, but these results indicate the presence of a wide proliferative Ki67 spectrum (18–89%), with patients in the p16 hetero/RB1+ group at the lowest end (mean percent of Ki67=42.5%; Figure 3b). These analyses have revealed key features of retinoblastoma pathway deregulation across the clinically distinct groups: CCNE1 gain/amplification in the p16 homo/RB1+ group, cyclin D1 overexpression in the p16 hetero/RB1+ group, and RB1 genomic alterations along with a high-proliferative index in the p16 homo/RB1− group.
p16/RB1 Deregulation Is an Early Event in High-Grade Serous Carcinoma Development
p16 and RB1 protein expression were analyzed by immunohistochemical analysis in the immediate precursor of high-grade serous carcinoma—serous tubal intraepithelial carcinoma, as recently defined by morphologic and immunohistochemistry criteria (p53 and Ki67; Supplementary Figure 2).20, 21, 22, 23, 24 Histological normal fimbriae (used as controls) showed weak p16 expression with few cells staining positive and intermediate levels of RB1 expression with dispersed regions of weaker staining (Supplementary Figure 3). p16 homogeneous expression was found in 73% (11/15) of serous tubal intraepithelial carcinomas, the same expression pattern as in their concomitant high-grade serous carcinoma, whereas only 20% (3/15) of these precursor lesions had p16 heterogeneous expression (Figure 4). The p16/RB1 subgroups identified in the established tumors were also found in serous tubal intraepithelial lesions at similar frequencies to the cancer: 47% of serous tubal intraepithelial carcinomas in the p16 homo/RB1+ subgroup, 27% p16 homo/RB1−, and 13% in the p16 hetero/RB1+ subgroup (Figure 4). Only one p16-/RB1+ serous tubal intraepithelial carcinoma was identified, a pattern shared with the corresponding high-grade serous carcinoma. This is in agreement with the high-grade serous carcinoma tumor data, which only had 23/334 p16- samples, suggesting that p16 inactivation is infrequent in this disease. These data suggest that retinoblastoma pathway deregulation occurs early in high-grade serous carcinoma tumorigenesis.
p16/RB1 immunohistochemistry patterns shared across serous tubal intraepithelial carcinomas and high-grade serous carcinoma ( × 20 magnification). (a) p16 homo/RB1+ subgroup identified in 47%. (b) p16 hetero/RB1+ subgroup identified in 13%. (c) p16 homo/RB1− subgroup identified in 27%. The prognostic signature identified in invasive cancers is also found in concomitant serous tubal intraepithelial carcinomas. The definition of serous tubal intraepithelial carcinoma includes morphological characteristics and immunohistochemical staining of Ki67 and p53.
Discussion
This study uses a large tumor cohort to comprehensively determine the different non-synonymous retinoblastoma pathway deregulations with relevance to patient outcome that predict for both progression-free survival as well as overall survival. Although, we were not able to identify any morphological differences among the 3 p16/Rb subgroups, in the cohort of 75 profiled tumors, a thorough investigation of morphological features on the entire set of 334 high-grade serous tumors is warranted.
We have also demonstrated that these clinically distinct subgroups are present in serous tubal intraepithelial carcinomas, the commonly accepted high-grade serous carcinoma precursor lesions, indicating that retinoblastoma pathway deregulation is an early event and that tumor behavior may be established early in ovarian tumorigenesis.
The striking p16 homogeneous expression pattern, frequently used as a marker of RB1 loss, seen in approximately 66% of high-grade serous carcinoma and uncommon in other epithelial ovarian cancer histotypes,7, 25, 26, 27, 28 predicts for a decreased progression-free survival and overall survival. The heterogeneous p16 pattern, present in about 33% of high-grade serous carcinoma cases and frequently seen in other epithelial ovarian cancer, predicts a better clinical outcome for high-grade serous carcinoma patients. Interestingly, loss of the CDKN2A gene is uncommon in high-grade serous carcinoma, found in only 6% (7/75) of our tumors. Copy number deletions in CDKN2A were infrequent and only three out of seven of those identified correlated with loss of protein expression, suggesting that retinoblastoma pathway dysfunction by CDKN2A loss occurs, but is an infrequent event in high-grade serous carcinoma.
The most common alteration associated with p16 homogeneous expression is loss of RB1. However, alterations in RB1 alone did not predict for clinical outcome. The observed loss of RB1 protein in approximately 30% of samples, correlated strongly with RB1 allelic loss and RB1 gene alterations (surveillance of deletions and mutations). RB1 inactivation through large spanning gene deletion occurred at a higher frequency (38%) than previously reported.5, 29 This is an interesting observation given the location of the RB1 gene on chromosome 13q14.2, a chromosomal arm not commonly reported as a deletion hotspot in high-grade serous carcinoma.5 Our mutation analysis and sequencing data are consistent with that of the TCGA research network results, showing infrequent RB1 point mutations in high-grade serous carcinoma.5, 30
Although high p16 is frequently used as a marker of RB1 loss, this is not the case in approximately 30% of our high-grade serous carcinoma samples, which show the shortest progression-free survival and shortest overall survival. The mechanism of p16 overexpression in the presence of functional RB1 is uncertain. Human papillomavirus infection, known to inactivate RB1 through binding to E7 proteins, was excluded by human papillomavirus testing (data not shown). There are few RB1 mutations known to maintain expression of the protein and few identified RB1 point mutations in high-grade serous carcinoma. Alternatively, alterations in CCNE1 may result in a bypass of the retinoblastoma pathway in this context, triggering p16 overexpression to inhibit continued proliferation.12 CCNE1 amplification is a known predictor of poor clinical outcome,31 but does not account for all of the cases in the p16 homo/RB1+ subgroup. Other yet unidentified oncogenes may also contribute to the retinoblastoma pathway deregulation in this particular subgroup.
Mechanisms of retinoblastoma pathway deregulation in other tumor types have previously been associated with clinical parameter differences including type, recurrence, and outcome,7, 8, 25, 26, 32, 33 and can be exploited for targeted therapy approaches to identify the most responsive patient subgroups.33, 34, 35 However, our study is unique because we have identified differential clinical behavior in a specific cancer histotype. This knowledge may have implications for identifying specific treatment strategies to improve care. Patients in the p16homo/RB1− subgroup show a significant increase in overall survival (>60 month; P=0.013), which suggests an increase in sensitivity to cytotoxic agents. In addition, this stratification could be used to determine novel second-line treatment options such as CDK4 inhibition (which inhibits the interaction with cyclin D1 resulting in hypophosphorylation of RB1 and E2F inactivation).15 Konecny et al. reported that ovarian cancer cell lines with RB1 deletions and CCNE1 gains are more resistant to CDK4 inhibitors, whereas those with CDKN2A gene deletions and decreased protein expression were more sensitive.15 We found that CDKN2A and/or CDK4 gene deletions are rare in high-grade serous carcinoma and that cyclin D1 overexpression is predominant in the p16 hetero/RB1+ tumors. Patients within the p16 hetero/RB1+ subgroup may be predicted to benefit most from CDK4 inhibitor treatments, in contrast to patients with cyclin E1 gains/amplifications in the p16 homo/RB1+ subgroup and RB1 deletions in the p16 homo/RB1− subgroup. Similarly, some high-grade serous carcinoma patients may optimally benefit from topoisomerase II inhibitors such as etoposide, which causes double-stranded DNA breaks and cell death by trapping topoisomerase proteins to DNA.36, 37, 38 Lung carcinoma studies indicate that RB1 mutations (found in 90% of small cell lung tumors) confer sensitivity to this treatment,9, 35, 39 suggesting that patients within the p16 homo/RB1− subgroup may also benefit from etoposide.
In summary, we have demonstrated by comprehensively assessing retinoblastoma pathway members: that epithelial ovarian cancer classified as high-grade serous carcinoma have non-synonymous mechanisms of retinoblastoma pathway deregulation, that these mechanisms result in different patterns of p16/RB1 protein expression, which stratify most high-grade serous carcinoma into three clinically relevant sub-groups, and that these alterations occur early in tumor development. Further studies in clinical trial settings would validate the utility of the retinoblastoma pathway subgroups in predicting the efficacy of novel targeted therapies.
References
Hennessy BT, Murph M, Nanjundan M et al. Ovarian cancer: linking genomics to new target discovery and molecular markers—the way ahead. Adv Exp Med Biol 2008;617:23–40.
Cho KR, Shih Ie M . Ovarian cancer. Annu Rev Pathol 2009;4:287–313.
Berns EM, Bowtell DD . The changing view of high-grade serous ovarian cancer. Cancer Res 2012;72:2701–2704.
Vaughan S, Coward JI, Bast RC Jr. et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer 2011;11:719–725.
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–615.
Tothill RW, Tinker AV, George J et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 2008;14:5198–5208.
Gauthier ML, Berman HK, Miller C et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 2007;12:479–491.
Loden M, Stighall M, Nielsen NH et al. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene 2002;21:4680–4690.
Zagorski WA, Knudsen ES, Reed MF . Retinoblastoma deficiency increases chemosensitivity in lung cancer. Cancer Res 2007;67:8264–8273.
Mohamed S, Yasufuku K, Hiroshima K et al. Prognostic implications of cell cycle-related proteins in primary resectable pathologic N2 nonsmall cell lung cancer. Cancer 2007;109:2506–2514.
Rayess H, Wang MB, Srivatsan ES . Cellular senescence and tumor suppressor gene p16. Int J Cancer 2012;130:1715–1725.
Witkiewicz AK, Knudsen KE, Dicker AP et al. The meaning of p16(ink4a) expression in tumors: functional significance, clinical associations and future developments. Cell Cycle 2011;10:2497–2503.
Kalof AN, Cooper K . p16INK4a immunoexpression: surrogate marker of high-risk HPV and high-grade cervical intraepithelial neoplasia. Adv Anat Pathol 2006;13:190–194.
Harbour JW, Dean DC . Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2000;2:E65–E67.
Konecny GE, Winterhoff B, Kolarova T et al. Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res 2011;17:1591–1602.
Richter S, Vandezande K, Chen N et al. Sensitive and efficient detection of RB1 gene mutations enhances care for families with retinoblastoma. Am J Hum Genet 2003;72:253–269.
Rushlow D, Piovesan B, Zhang K et al. Detection of mosaic RB1 mutations in families with retinoblastoma. Hum Mutat 2009;30:842–851.
Zeschnigk M, Lohmann D, Horsthemke B . A PCR test for the detection of hypermethylated alleles at the retinoblastoma locus. J Med Genet 1999;36:793–794.
Banks RE, Tirukonda P, Taylor C et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res 2006;66:2000–2011.
Shaw PA, Rouzbahman M, Pizer ES et al. Candidate serous cancer precursors in fallopian tube epithelium of BRCA1/2 mutation carriers. Mod Pathol 2009;22:1133–1138.
Visvanathan K, Vang R, Shaw P et al. Diagnosis of serous tubal intraepithelial carcinoma based on morphologic and immunohistochemical features: a reproducibility study. Am J Surg Pathol 2011;35:1766–1775.
Jarboe E, Folkins A, Nucci MR et al. Serous carcinogenesis in the fallopian tube: a descriptive classification. Int J Gynecol Pathol 2008;27:1–9.
Lee Y, Miron A, Drapkin R et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol 2007;1:26–35.
Vang R, Visvanathan K, Gross A et al. Validation of an algorithm for the diagnosis of serous tubal intraepithelial carcinoma. Int J Gynecol Pathol 2012;31:243–253.
Romagosa C, Simonetti S, Lopez-Vicente L et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 2011;30:2087–2097.
Berman HK, Gauthier ML, Tlsty TD . Premalignant breast neoplasia: a paradigm of interlesional and intralesional molecular heterogeneity and its biological and clinical ramifications. Cancer Prev Res (Phila) 2010;3:579–587.
Yemelyanova A, Ji H, Shih Ie M et al. Utility of p16 expression for distinction of uterine serous carcinomas from endometrial endometrioid and endocervical adenocarcinomas: immunohistochemical analysis of 201 cases. Am J Surg Pathol 2009;33:1504–1514.
O'Neill CJ, McBride HA, Connolly LE et al. High-grade ovarian serous carcinoma exhibits significantly higher p16 expression than low-grade serous carcinoma and serous borderline tumour. Histopathology 2007;50:773–779.
Gras E, Pons C, Machin P et al. Loss of heterozygosity at the RB-1 locus and pRB immunostaining in epithelial ovarian tumors: a molecular, immunohistochemical, and clinicopathologic study. Int J Gynecol Pathol 2001;20:335–340.
Kuo KT, Guan B, Feng Y et al. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res 2009;69:4036–4042.
Etemadmoghadam D, George J, Cowin PA et al. Amplicon-dependent CCNE1 expression is critical for clonogenic survival after cisplatin treatment and is correlated with 20q11 gain in ovarian cancer. PLoS One 2010;5:e15498.
Herschkowitz JI, He X, Fan C et al. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res 2008;10:R75.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70.
Dean JL, McClendon AK, Hickey TE et al. Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors. Cell Cycle 2012;11:2756–2761.
Brambilla E, Gazdar A . Pathogenesis of lung cancer signalling pathways: roadmap for therapies. Eur Respir J 2009;33:1485–1497.
Treszezamsky AD, Kachnic LA, Feng Z et al. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res 2007;67:7078–7081.
Kingma PS, Burden DA, Osheroff N . Binding of etoposide to topoisomerase II in the absence of DNA: decreased affinity as a mechanism of drug resistance. Biochemistry 1999;38:3457–3461.
van Maanen JM, Retel J, de Vries J et al. Mechanism of action of antitumor drug etoposide: a review. J Natl Cancer Inst 1988;80:1526–1533.
Brambilla E, Moro D, Gazzeri S et al. Alterations of expression of Rb, p16(INK4A) and cyclin D1 in non-small cell lung carcinoma and their clinical significance. J Pathol 1999;188:351–360.
Acknowledgements
We thank the UHN Biobank for sample acquisition, Diane Rushlow from Impact Solutions for RB1 mutation oversight, Alice Chun, the TCAG for performing the SNP microarrays, and the UHN Pathology Research Program and James Ho at the AMPL, for immunohistochemistry services. We also thank the Gynecological Oncologists at UHN for enabling sample access. This study was funded by the CDMRP Ovarian Cancer Research program (DOD W81WH-0701-0371), the PMCF Appel Fund, and the OGS fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Milea, A., George, S., Matevski, D. et al. Retinoblastoma pathway deregulatory mechanisms determine clinical outcome in high-grade serous ovarian carcinoma. Mod Pathol 27, 991–1001 (2014). https://doi.org/10.1038/modpathol.2013.218
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.218
Keywords
This article is cited by
-
A comprehensive immunohistochemical analysis of 26 markers in 250 cases of serous ovarian tumors
Diagnostic Pathology (2023)
-
Genomic profiling in ovarian cancer retreated with platinum based chemotherapy presented homologous recombination deficiency and copy number imbalances of CCNE1 and RB1 genes
BMC Cancer (2019)
-
Loss of LKB1 and p53 synergizes to alter fallopian tube epithelial phenotype and high-grade serous tumorigenesis
Oncogene (2016)