Abstract
Idiopathic pulmonary fibrosis is a fatal disease without effective therapy or diagnostic test. To investigate a potential role for γ−herpesviruses in this disease, 21 paraffin-embedded lung biopsies from patients diagnosed with idiopathic pulmonary fibrosis and 21 lung biopsies from age-matched controls with pulmonary fibrosis of known etiology were examined for a series of γ−herpesviruses’ DNA/RNA and related proteins using in situ hybridization and reverse transcriptase-polymerase chain reaction (RT-PCR)-based methods. We detected four proteins known to be in the genome of several γ−herpesviruses (cyclin D, thymidylate synthase, dihydrofolate reductase, and interleukin-17) that were strongly co-expressed in the regenerating epithelial cells of each of the 21 idiopathic pulmonary fibrosis cases and not in the benign epithelia of the controls. Among the γ−herpesviruses, only herpesvirus saimiri expresses all four of these ‘pirated’ mammalian proteins. We found herpesvirus saimiri DNA in the regenerating epithelial cells of 21/21 idiopathic pulmonary fibrosis cases using four separate probe sets but not in the 21 controls. RT-PCR showed that the source of the cyclin D RNA in active idiopathic pulmonary fibrosis was herpesvirus saimiri and not human. We cloned and sequenced part of genome corresponding to the DNA polymerase herpesvirus saimiri gene from an idiopathic pulmonary fibrosis sample and it matched 100% with the published viral sequence. These data are consistent with idiopathic pulmonary fibrosis representing herpesvirus saimiri-induced pulmonary fibrosis. Thus, treatment directed against viral proliferation and/or viral-associated proteins may halt disease progression. Further, demonstration of the viral nucleic acids or proteins may help diagnose the disease.
Similar content being viewed by others
Main
The (gamma) γ-herpesvirus family is a candidate for causing idiopathic pulmonary fibrosis in humans. Specifically, γ-herpesviruses cause pulmonary fibrosis in horses (equine herpesvirus 5)1 and donkeys (asinine herpesvirus)2 that clinically resemble idiopathic pulmonary fibrosis in humans. Murine herpesvirus-68 (MHV68),3 a γ-herpesvirus that is closely related to herpesvirus saimiri, is used to cause pulmonary fibrosis in mice4 that was successfully arrested with antiviral therapy;5 this is the only recognized viral animal model of idiopathic pulmonary fibrosis.
Herpesvirus saimiri is a γ2-herpesvirus that is non-pathogenic in its natural host, the squirrel monkey (Saimiri sciureus), but causes fatal T-cell lymphomas and leukemias in New World monkeys.6 In vitro, it infects a wide variety of human cell types7, 8, 9 and transforms Rat-1 cells to form foci in culture.10 Subclinical infections in humans may be common, as an estimated 4.0–7.3% of people are seropositive for herpesvirus saimiri.11 Productive infection by herpesvirus saimiri is associated with the co-expression of viral DNA with several pirated mammalian proteins such as interleukin-17 (IL-17),12, 13 viral cyclin D,14 thymidylate synthase,15 and dihydrofolate reductase.16
About 200 000 in the United States of America have idiopathic pulmonary fibrosis, which is typically fatal within 2 years of diagnosis and with no diagnostic test.17 Idiopathic pulmonary fibrosis does not respond to combined immunosuppressive and systemic steroid therapy, which may worsen the symptomatology.18 Other common treatment combinations have also failed to show efficacy in multicenter clinical trials.19 The theoretical possibility of infectious etiology for idiopathic pulmonary fibrosis is currently being re-examined.20
Our recent work revealed the unique role that regenerating epithelial cells have in idiopathic pulmonary fibrosis.21 These cells are the primary sources of a variety of immunomodulating proteins in idiopathic pulmonary fibrosis, including IL-17, a cytokine that was discovered in activated T cells,13 with high homology to a gene in herpesvirus saimiri.12 Here, we report a likely etiologic role of herpesvirus saimiri in idiopathic pulmonary fibrosis in humans by showing a strong co-expression of IL-17, cyclin D1, thymidylate synthase, and dihydrofolate reductase with the viral DNA. In pulmonary fibrosis of known etiology, there is lack of herpesvirus saimiri DNA, and there is an absence of co-expression of herpesvirus saimiri-pirated proteins in a large series of benign and malignant diseases of the lung. We also report the demonstration by RT-PCR that the cyclin D in idiopathic pulmonary fibrosis is expressed from the corresponding gene of herpesvirus saimiri and not from the human host, and the cloning and sequencing of a part of the viral DNA polymerase gene from a clinical sample of idiopathic pulmonary fibrosis that revealed a 100% match with the published sequence of herpesvirus saimiri.22
Materials and methods
Patient Samples
The patient samples (formalin-fixed biopsies) from the files of Weill Cornell Medical Center or the Medical College of Wisconsin had an IRB-approved protocol that de-identified all data except for the diagnosis, age, extent of negative workup, and sex of the patient. Twenty-one cases of idiopathic pulmonary fibrosis were obtained and were diagnosed as such after review by clinicians, radiologists, and pathologists (SS, CM) expert in the field. The American Thoracic Society and the European Respiratory Society criteria of idiopathic pulmonary fibrosis were used that included a histologic diagnosis of usual interstitial pneumonitis, abnormal pulmonary function tests indicative of restrictive lung disease, abnormal findings on CT scan, age >50 years, duration of illness >3 months, and exclusion of known causes of interstitial lung disease. The mean age of the patients was 60.6 years (s.e.m.=2.3 years); 14 were men and 7 were women. As controls, 21 paraffin-embedded biopsies of focal lung fibrosis of known cause were studied (mean age 62.0 with 15 men). These included seven cases of fibrosis-related adenocarcinoma of the lung, five cases of lung fibrosis associated with emphysema, and nine cases of interstitial pneumonitis and fibrosis of known viral etiology including measles (one case), adenovirus (three cases), hantavirus (three cases), and rotavirus (two cases). The lung fibrosis associated with emphysema was chosen to show only focal fibrosis, with no evidence of co-existing lung disease such as desquamative interstitial pneumonia or usual interstitial pneumonia. The cases and controls were all from open lung biopsies. As additional negative and positive controls, we infected Jurkat cells with herpesvirus saimiri obtained from the ATCC (catalog no. F28513, Manassas VA, USA). Three days post transfection, the cells were fixed in 10% buffered formalin for 5 h and then processed for in situ hybridization.
In Situ Hybridization Analysis
Our in situ hybridization protocol has been previously described.23 In brief, in situ hybridization was performed on each of the 21 cases and 21 controls for herpesvirus saimiri. The probes were LNA (locked nucleic acid)-modified and 5′ digoxigenin-tagged (Exiqon). Two separate probe sets against herpesvirus saimiri were tested: probe set one was directed against ORF1, the herpesvirus Saimiri Transforming Protein (STP) oncogene and had the following sequences: 5′-CTCTAAGCACAGGGGCACAG-3′ and 5′-CTACGCAGAAGTCGGAAGCC-3′.24 The second probe set was directed against the 1444-base pair repeat, heavy-DNA (H-DNA) non-transcribed region, which is present in about 35 copies/viral genome and had the following sequences: 5′-GCCGCCTCAGAATTTTAGCA-3′ and 5′-CTCTGCGTGAAGCACAGTGC-3′.25 These in situ hybridization tests were performed with serial sections to compare the same groups of cells. The majority of the cases were also tested by in situ hybridization for Epstein-Barr virus (EBV), cytomegalovirus (CMV), and herpes simplex virus types I/II (HSVI/II) using biotin-labeled probes from Enzo Life Sciences. The probe/target complex was visualized after the alkaline phosphatase-linked conjugate reacted with the chromogen, nitroblue tetrazolium, and bromochloroindolyl phosphate (NBT/BCIP) with a nuclear fast red counterstain. Negative controls included omission of the probe, use of a scrambled probe, non-infected Jurkat cells, and the 21 cases of lung fibrosis of known etiology.
Immunohistochemical Analysis
Our immunohistochemical protocol has been previously described (Leica BOND-MAX).23 We analyzed for the latent membrane protein (LMP) of EBV, the latent nuclear antigen (LNA-1) of Kaposi’s associated herpesvirus (KSHV), and CMV proteins 8B1.2, 1G5.2, and 2D4.2. As productive infection by herpesvirus saimiri is associated with the expression of several pirated mammalian proteins, including viral cyclin D, thymidylate synthase, IL-17, and dihydrofolate reductase, and as each of these four proteins shares sufficient homology with the counterpart human protein to be detected using immunohistochemistry,6 we also analyzed the cases and the controls for each of these four proteins.
Co-expression Analysis
Our co-expression analysis protocol has been previously reported.21, 23 The computer-based analysis by the Nuance system (Caliper) separates each chromogenic spectral signal, converts it to a fluorescent signal, then mixes the two and indicates whether cells contain the two targets of interest.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from paraffin samples using the RecoverAll Total Nucleic Acid Isolation Kit (AM1975, Ambion) according to the manufacturer’s instructions. cDNA was generated from this RNA using SuperScript III First-Strand Synthesis SuperMix Kit (18080400, Invitrogen) using the following primers:
Human Cyclin D1 5′-CGGAGGAGAACAAACAGATCATCCGCAAAC-3′, 5′-GTGTGAGGCGGTAGTAGGACAGGAAGTTGT-3′; Viral Cyclin D 5′-ACTGCTTACCTGGATGCATCTGCTCTGTGA-3′, 5′-GCAAGTACAGCTTCAGTGTGTCCCATTTCAGTGC-3′. The viral and human primers share no homology. PCR was conducted with Ex Taq DNA Polymerase (RR01A, Takara) and the primers are listed above.
Results
Initially, we addressed the question of whether idiopathic pulmonary fibrosis was associated with any herpesvirus infection. We screened 13 cases of idiopathic pulmonary fibrosis via in situ molecular pathology analyses for different γ-herpesviruses and HSV I/II. Each case was negative for EBV, CMV, Kaposi’s sarcoma herpes virus, and HSV I/II. Each of the 13 idiopathic pulmonary fibrosis cases was positive for herpesvirus saimiri DNA. We expanded the study to a total of 21 cases of idiopathic pulmonary fibrosis and 21 cases of age-matched controls, which showed pulmonary fibrosis of known etiology. When read blinded to the diagnosis, 21/21 of idiopathic pulmonary fibrosis cases were positive for herpesvirus saimiri DNA by in situ hybridization, whereas 0/21 of the control cases of pulmonary fibrosis was herpesvirus saimiri DNA-positive. Further, the Jurkat cells infected by the virus showed an intense nuclear signal in the majority of the malignant cells, whereas the non-infected cells had no signal (data not shown).
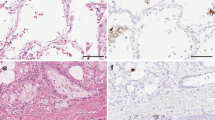
The herpesvirus saimiri DNA distribution closely paralleled the histopathology of usual interstitial pneumonitis that is evident in idiopathic pulmonary fibrosis (Figures 1a–c). Herpesvirus saimiri-positive nuclei were evident in many of the regenerating epithelial cells in the areas of active and early fibrosis (Figure 1d, g and h). Although the histologically normal lung in idiopathic pulmonary fibrosis was often viral-negative, rare viral DNA-positive pneumocytes were at times evident (Figure 1e). Virus-positive cells were not evident in the areas of end-stage fibrosis of idiopathic pulmonary fibrosis, which lacked epithelial cells, or in the regenerating epithelial cells of interstitial pneumonitis with fibrosis of known etiology (Figure 1f). High magnification (Figure 1c) showed that the herpesvirus saimiri-positive epithelial cells often showed nuclear atypia and, at times, multinucleation.
Histopathological and molecular correlates of idiopathic pulmonary fibrosis. a–c (hematoxylin and eosin) show increased magnifications of idiopathic pulmonary fibrosis in which areas of histologically normal lung is admixed with areas of active fibrosis (a, × 25), marked by serpentine glands lined with epithelia (b, × 50), which show nuclear atypia and multinucleation (c, arrow, × 400). Herpesvirus saimiri DNA was commonly detected in the nuclei of the epithelia of these serpentine glands (d, × 50) and much less evident in the admixed histologically normal lung (e, × 400; blue NBT/BCIP signal with pink counterstain). In comparison, herpesvirus saimiri DNA was not evident in cases of interstitial pneumonitis and fibrosis of known viral etiology (f, × 400 measles infection). Herpesvirus saimiri DNA was evident in the regenerating epithelia in idiopathic pulmonary fibrosis in the same cells when serial sections were probed for herpesvirus saimiri terminal repeat sequence (g, × 400) or the STP ORF 1 gene (h, × 400).
The different γ-herpesvirus members show a similar genetic organization and share homology over various parts of their genomes.6, 22, 26, 27 The probe set used for the initial evaluation used two probes for the herpesvirus saimiri-specific oncogene, STP. A BLAST sequence analysis showed that probe 1 shared weak homology with some human genes, whereas probe 2 had no such homology. In situ hybridization was performed on serial sections using probe 1 and probe 2 separately in five cases. In each case, STP probe 1 and STP probe 2 resulted in a signal in the same group of regenerating epithelial cells (data not shown). Further, we separated the two LNA probes targeting the 1444-base pair DNA repeat region of herpesvirus saimiri (H-DNA region) and analyzed the serial sections of another set of five cases of idiopathic pulmonary fibrosis. In each case, individual probes corresponding to the H-DNA viral region yielded signals in the same groups of regenerating epithelial cells as the probe cocktail containing both H-DNA probes 1 and 2 (data not shown). Similarly, testing serial sections of idiopathic pulmonary fibrosis with the H-DNA and STP probe sets showed the same viral distribution in the adjacent sections (Figure 1, panels G and H, respectively).
The data showing a strong nuclear-based signal in the proposed target cell of idiopathic pulmonary fibrosis are consistent with a productive viral infection, as any herpes virus infection is invariably nuclear-based. To seek additional evidence, we analyzed serial sections of the cases of idiopathic pulmonary fibrosis that had sufficient tissue for known herpesvirus saimiri-pirated mammalian proteins including IL-17, cyclin D, thymidylate synthase, and dihydrofolate reductase with matching negative controls. When examined blinded to the diagnosis, in each of the idiopathic pulmonary fibrosis cases, there was marked overexpression of cyclin D (Figures 2a and b), dihydrofolate reductase (Figure 2c), thymidylate synthase, and IL-17. Note the equivalent distribution patterns for the viral-associated proteins and herpesvirus saimiri DNA as they localized to the regenerating epithelial cells in areas of active fibrosis. Further, the proteins were much less evident in the epithelia of the histologically unremarkable lung in the idiopathic pulmonary fibrosis cases (Figure 2d).
Expression of herpesvirus saimiri-related proteins in idiopathic pulmonary fibrosis. a ( × 100, fast red stain and hematoxylin counterstain) and, at higher magnification, b ( × 400) show that cyclin D1 is highly expressed in the regenerating epithelial cells of idiopathic pulmonary fibrosis. c ( × 100) shows the same pattern of localization for dihydrofolate reductase to the regenerating epithelia in the areas of active fibrosis of idiopathic pulmonary fibrosis and the rarity of the protein in the adjacent normal areas of the same lung tissue (d, × 200). The tissues in e–h (each at × 400) represent photographs of the same cells in serial sections of a case of idiopathic pulmonary fibrosis. The samples were analyzed for either herpesvirus saimiri using an probe specific for the IL-17 DNA sequence (e, f) or the herpesvirus saimiri DNA polymerase gene (g, h). After in situ hybridization, co-expression analysis was performed using immunohistochemistry with either cyclin D (e, f) or dihydrofolate reductase (g, h). In each case, the signal for the DNA was blue and the protein was brown; the Nuance-merged image shows the DNA as fluorescent blue and the protein as fluorescent green, whereas cells with both targets show fluorescent yellow. Note that the same pattern of co-expression is seen in e–h in the merged images.
The protein and herpesvirus saimiri DNA data are presented in Table 1. Note that expression levels of cyclin D1, thymidylate synthase, and dihydrofolate reductase were much less evident in interstitial pneumonitis and fibrosis of known viral etiology or in the fibrosis associated with emphysema or adenocarcinoma when compared with idiopathic pulmonary fibrosis. Although these proteins were commonly detected in the adenocarcinoma cells, they were rarely present in the regenerating lung epithelia and desmoplasia present at the tumor-benign lung interface (data not shown). Importantly, none of the controls had any of the four proteins co-expressed by the same cells. Also note that Table 1 includes the serial section analysis of 41 benign stromal tumors, 34 sarcomas, and 44 non-small cell carcinomas of the lung. Note that the majority of the lung cancers and sarcomas were associated with dihydrofolate reductase expression. However, not one sarcoma, carcinoma, or control case demonstrated co-expression for dihydrofolate reductase and cyclin D or IL-17. Thus, the co-expression of dihydrofolate reductase, cyclin D, thymidylate synthase, and IL-17 proteins seemed to be unique to idiopathic pulmonary fibrosis.
We cloned parts of the herpesvirus saimiri genome corresponding to the DNA polymerase and IL-17 viral genes, given that the former is well conserved among the herpesvirus saimiri strains and the latter is specific for herpesvirus saimiri. These clones were used to generate biotin-labeled probes that were tested on selected cases of idiopathic pulmonary fibrosis and matching controls. When tested in serial sections, the probes for the STP, IL-17, DNA polymerase, and terminal repeat viral sequences yielded signals in the same regenerating epithelial cells in idiopathic pulmonary fibrosis (Figures 2e–h) and not in the negative controls. Co-expression analysis also showed that the same sets of cells that were expressing these viral DNA sequences co-expressed cyclin D (Figures 2e–h, respectively).
To obtain further evidence of the strong co-expression in idiopathic pulmonary fibrosis, we performed direct co-expression analysis of the viral DNA with dihydrofolate reductase, cyclin D, thymidylate synthase, and IL-17 in the 18 idiopathic pulmonary fibrosis cases where sufficient tissue remained. As seen in Figure 3, co-expression analysis confirmed that epithelial cells in the active areas of idiopathic pulmonary fibrosis that were positive for viral DNA were also co-expressing the herpesvirus saimiri-pirated proteins (Figures 3a and b, herpesvirus saimiri DNA and thymidylate synthase; Figures 3c and d, herpesvirus saimiri DNA and IL-17; Figure 3e and f cyclin D1 and herpesvirus saimiri DNA (STP probe set); and Figure 3g and h, cyclin D1 and herpesvirus saimiri DNA, H-DNA probe set). We also had positive co-expression for herpesvirus saimiri and cytokeratin AE1/3 (a marker of regenerating epithelial cells), demonstrating that essentially all of the viral DNA-positive cells were indeed cytokeratin-positive (data not shown). Finally, co-expression analysis of cyclin D1 with thymidylate synthase, IL-17, and dihydrofolate reductase confirmed that basically all of the regenerating epithelial cells expressing cyclin D1 were also expressing the other three viral-related proteins (Figure 4).
Co-expression analysis of herpesvirus saimiri DNA and related proteins in idiopathic pulmonary fibrosis. Each set of panels shows the RGB image (a, c, e, and g) and the Nuance-derived image (b, d, f, and h) after co-expression analysis of viral DNA (blue in the RGB image and fluorescent blue in the Nuance image) and the protein (DAB for a and c and fast red for e and g; each protein is fluorescent red in the Nuance image). Fluorescent yellow in b, d, f, and h reflects co-expression of the viral DNA and the protein; each image is at × 400. Herpesvirus saimiri DNA strongly co-expressed with thymidylate synthase (a, b), IL-17 (c and d), and cyclin D1 (e and f). e/f and g/h are serial sections in which the same cells were analyzed for co-expression for cyclin D1 and the herpesvirus saimiri DNA STP gene (e and f) and the terminal repeat sequence (g and h), respectively.
Detection of viral cyclin RNA using RT-PCR analysis in idiopathic pulmonary fibrosis. In a, it is evident that samples 2392 (negative for idiopathic pulmonary fibrosis) and 21 565 (histologically normal area of idiopathic pulmonary fibrosis case with no detectable viral DNA) showed only human cyclin D1 cDNA amplicons after RT-PCR (viral amplicon 290 nucleotides and human amplicon 180 nucleotides). In comparison, the RNA extracted from two cases of idiopathic pulmonary fibrosis with active fibrosis and 2+ viral DNA (39B1 and 51-1) showed primarily viral cyclin D1 amplicons after RT-PCR analysis. b shows that in an idiopathic pulmonary fibrosis case with 3+ herpesvirus saimiri DNA (2210), only viral cyclin D RNA sequence is amplified and that the band intensity is greater than that for the 2+ viral cases shown in a. c shows the strong expression of cyclin D using immunohistochemistry in case 2210 plus the strong co-expression of cyclin D1 (fluorescent green) and dihydrofolate reductase (fluorescent red) with the merged image showing co-expression as fluorescent yellow.
We next extracted the RNA from five samples to determine whether the cyclin D RNA that was highly expressed in idiopathic pulmonary fibrosis was viral, human, or both in origin. We were limited to five samples as we only had formalin-fixed, paraffin-embedded tissues in each control and each idiopathic pulmonary fibrosis case, except for one case where frozen unfixed tissue was available. For many of the samples, we used most of the block for the in situ-based analyses and only five yielded sufficient RNA for RT-PCR. We chose to study cyclin D RNA, as it has the least homology between human and viral genes (23%) of the four pirated genes, making it straightforward to design primers with no homology between the corresponding human and viral cyclin RNA sequences. The viral-specific amplicons derived from RTPCR were 290 nucleotides, whereas the human-specific amplicons were 180 nucleotides. When analyzed blindly (Figure 4), the case with no idiopathic pulmonary fibrosis (normal lung adjacent to cancer, lane 2392) showed a human cyclin D1 cDNA band and no viral band. Case 21565, which was a histologically normal lung from a case of idiopathic pulmonary fibrosis that showed no viral DNA on in situ hybridization, also showed a human cyclin D1 and no viral band. The two cases of active idiopathic pulmonary fibrosis with a 2+ viral copy number (39B and 51-1) showed primarily viral cyclin D cDNA amplicons. The one case of active idiopathic pulmonary fibrosis that showed 3+ viral DNA with 3+ expression of cyclin D upon immunohistochemistry (Figure 4) showed the most intense band with the viral-specific cyclin D primers and no band with the human-specific cyclin D1 primers. Thus, there was a good correlation between the in situ findings of herpesvirus saimiri DNA detection and the viral origin of the cyclin D RNA.
Finally, we used the DNA extracted from the frozen, fixed tissue of idiopathic pulmonary fibrosis in PCR analysis to amplify part of the herpesvirus saimiri DNA polymerase gene. An H&E-stained section of the tissue from the case confirmed both idiopathic pulmonary fibrosis and many regenerating epithelial cells positive for the virus DNA and viral-related proteins (Figure 4, panel C). After amplifying the DNA, we cloned and sequenced it. The sequence of the amplicon internal to the primers (5′-GGGTGCGAGCGTATGTGTTAACGTGTTTGGACAAAGAAATTACTTTTATGT-3′) was a 100% match with the reported sequence of herpesvirus saimiri strain C488 and shows only one-base pair mismatch with strain A11 of herpesvirus saimiri,22 as would be expected when sequencing a well-conserved region of herpesvirus saimiri.
Discussion
A primary finding of this study is that thymidylate synthase, dihydrofolate reductase, IL-17, and a viral cyclin D1 homolog are strongly co-expressed in the regenerating epithelial cells that are orchestrating the pathophysiology of idiopathic pulmonary fibrosis and not in the epithelia of non-idiopathic pulmonary fibroses including multiple cases of interstitial pneumonitis and fibrosis of known viral etiology. Among the γ-herpesviruses, the combination of these proteins is unique to herpesvirus saimiri. We have shown that herpesvirus saimiri DNA, using four separate probes corresponding to different regions of the viral genome, and each of these four pirated mammalian proteins, co-localized to the regenerating epithelial cells of idiopathic pulmonary fibrosis. We chose to examine the cyclin D RNA as the sequences are most disparate when comparing viral to human sequences. RT-PCR demonstrated that the primary source of cyclin D in active idiopathic pulmonary fibrosis is from herpesvirus saimiri and not from the human host. We were able to clone the DNA from a patient’s idiopathic pulmonary fibrosis sample using PCR and primers from the viral DNA polymerase chain gene. When the clone was sequenced, it revealed a perfect match with the reported sequence for herpesvirus saimiri.
The data suggest that the detection of these proteins and viral DNA in a patient with pulmonary fibrosis would be diagnostic of idiopathic pulmonary fibrosis and, importantly, may help in dictating effective therapy. In this regard, there are reports in both human idiopathic pulmonary fibrosis28 and the murine model of PF,5, 29 in which anti-herpesvirus treatment was effective in halting disease progression. Further, rapamycin therapy, which inhibits cyclin D1 activity,30 has been shown to stop disease progression in a human case of idiopathic pulmonary fibrosis31 and in a murine model of this disease.32
Herpesvirus saimiri is a member of the γ-herpesvirus subfamily that shares the same genetic organization as other members that are strongly associated with human disease, including EBV, CMV, and Kaposi’s sarcoma herpes virus.6 The genomes of these viruses are very large (∼160 kb) and are arranged in a series of ORFs that comprise 83 potential genes. Although all herpesvirus family members have incorporated DNA sequences that share homology with mammalian sequences, herpesvirus saimiri is notable for having the largest number of pirated human and mammalian DNA sequences of any known virus.22 Little is known about the consequences of herpesvirus saimiri infection in humans. The older age (>50 years is a minor criterion of idiopathic pulmonary fibrosis with the American Thoracic Society and the European Respiratory Society criteria) of patients diagnosed with idiopathic pulmonary fibrosis33 suggests that reactivation of latent herpesvirus infection may be occurring as in the well-documented occurrence with varicella-zoster and shingles.
The histopathology of idiopathic pulmonary fibrosis is marked by many microscopic areas of alveolar damage with regenerating epithelial cells and early fibrosis interspersed with histologically normal lung and end-stage fibrosis with the loss of epithelia. This heterogeneous pattern, although typical of idiopathic pulmonary fibrosis, is not diagnostic because it can overlap with other causes of interstitial pneumonitis, including those caused by a variety of acute viral infections and various drugs. Further, the histologic pattern of idiopathic pulmonary fibrosis is consistent with a multifocal pulmonary infectious process with a temporally heterogeneous shift from latent to productive infection in many different foci. The in situ methods used in this study demonstrated a parallel viral pattern with rare cells positive for viral DNA as well as the viral-associated pirated proteins in the normal areas of idiopathic pulmonary fibrosis, a marked increase in viral DNA and the pirated proteins in the active areas of idiopathic pulmonary fibrosis, and the loss of viral DNA and associated proteins in the honeycombed areas of end-stage fibrosis.
In sum, these data show that idiopathic pulmonary fibrosis is associated with strong co-expression of four herpesvirus saimiri-associated proteins, which were not found in aggregate in either benign or cancerous lung tissues or in a large series of sarcomas and that these proteins co-localize with herpesvirus saimiri DNA. Although the virus is likely herpesvirus saimiri, we cannot exclude the possibility of a different thus far undiscovered γ-herpesvirus closely related to herpesvirus saimiri that shares the DNA and RNA sequences tested in this project. However, this seems unlikely, as the PCR-obtained clone from a patient’s idiopathic pulmonary fibrosis sample showed 100% homology with the reported sequence of the herpesvirus saimiri DNA polymerase. Given that idiopathic pulmonary fibrosis has no effective treatment or specific diagnostic test and is typically fatal within several years of diagnosis,33 these data may provide a diagnostic test for this disease and offer patients the hope of stabilization of their disease via treatments directed against herpesvirus saimiri proliferation.
References
Williams KJ, Maes R, Piero FD et al. Equine multinodular pulmonary fibrosis: a newly recognized herpesvirus-associated fibrotic lung disease. Vet Path 2007;44:849–862.
Kleiboeker SB, Schommer SK, Johnson PJ et al. Association of two newly recognized herpesviruses with interstitial pneumonia in donkeys (Equus asinus). J Vet Diagn Invest 2002;14:273–280.
Virgin HW, LaTreille P, Wamsley P et al. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 1997;71:5894–5904.
Mora AL, Woods CR, Garcia A et al. Lung infection with gamma-herpesvirus induces progressive pulmonary fibrosis in Th2-biased mice. Am J Physiol Lung Cell Mol Physiol 2005;289:L711–L721.
Mora AL, Torres-Gonzales E, Rojas M et al. Control of virus reactivation arrests pulmonary herpesvirus-induced fibrosis in IFN-g receptor-deficient mice. Am J Respir Crit Care Med 2007;175:1139–1150.
Fickenscher H, Fleckenstein B . Herpesvirus saimiri. Phil Trans Royal Soc London B 2001;356:545–567.
Simmer B, Alt M, Buckreus I et al. Persistence of selectable herpesvirus saimiri in various human haematopoietic and epithelial cell lines. J Gen Virol 1991;72:1953–1958.
Biesinger B, Muller-Fleckenstein I, Simmer B et al. Stable growth transformation of human T lymphocytes by Herpesvirus saimiri. Proc Natl Acad Sci USA 1992;89:3116–3119.
Smith PG, Burchill SA, Brooke D et al. Efficient infection and persistence of a herpesvirus saimiri-based gene delivery vector into human tumor xenographs and multicellular spheroid cultures. Cancer Gene Ther 2005;12:248–256.
Jung JU, Trimble JJ, King NW et al. Identification of transforming genes of subgroup A and C strains of Herpesvirus Saimiri. Proc Natl Acad Sci USA 1991;88:7051–7055.
Ablashi D, Dahlberg J, Cannon G et al. Detection of antibodies to Herpesvirus saimiri late antigens in human sera. Intervirol 1988;29:217–226.
Yao Z, Fanslow WC, Seldin MF et al. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity 1995;3:811–821.
Rouvier E, Luciani M-F, Mattei M-G et al. CTLA-8 cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a Herpesvirus Saimiri gene. J Immunol 1993;150:5445–5456.
Nicholas J, Cameron KR, Honess RW . Herpesvirus saimiri encodes homologues of G-protein-coupled receptors and cyclins. Nature 1992;355:362–365.
Bodemer W, Niller HH, Nitsche N et al. Organization of the thymidylate synthase gene of herpesvirus saimiri. J Virol 1986;60:114–123.
Trimble JJ, Murthy SCS, Bakker A et al. A gene for dihydrofolate reductase in a herpesvirus. Science 1988;239:1145–1147.
Olsen AL, Swigris JJ, Lezotte DC et al. Mortality from pulmonary fibrosis increased in the United States from 1992 to 2003. Am J Respir Crit Care Med 2007;176:277–284.
Papiris SA, Manali ED, Kolilekas L et al. Steroids in idiopathic pulmonary fibrosis acute exacerbation: defenders or killers? Am J Respir Crit Care Med 2012;185:587–588.
IPFCRN. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med 2012;366:1968–1977.
Cookson WOC, Moffatt MF . Bedside to gene and back in idiopathic pulmonary fibrosis. N Engl J Med 2013;368:2228–2230.
Nuovo GJ, Hagood JS, Magro CM et al. The distribution of immunomodulatory cells in the lungs of patients with idiopathic pulmonary fibrosis. Mod Pathol 2012;25:416–433.
Albrecht J-C, Nicholas J, Biller D et al. Primary structure of the herpesvirus saimiri genome. J Virol 1992;66:5047–5058.
Nuovo GJ . In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods 2010;52:307–315.
Yasukawa M, Inoue Y, Kimura N et al. Immortalization of human T cells expressing T-cell receptor gamma-delta by Herpesvirus Saimiri. J Virol 1995;69:8114–8117.
Bankier AT, Dietrich W, Baer R et al. Terminal repetetive sequences in herpesvirus saimiri virion DNA. J Virol 1985;55:133–139.
Jung JU, Stager M, Desrosiers RC . Virus-encoded cyclin. Mol Cell Biol 1994;14:7235–7244.
Honess RW, Bodemer W, Cameron KR et al. The A+T-rich genome of herpesvirus saimiri contains a highly conserved gene for thymidylate synthase. P Proc Natl Acad Sci USA 1986;83:3604–3608.
Tang Y-W, Johnson JE, Browning PJ et al. Herpesvirus DNA is consistently detected in lungs of patients with idiopathic pulmonary fibrosis. J Clin Microbiol 2003;41:2633–2640.
Neyts J, De Clercq E . In vitro and in vivo inhibition of Murine Gamma Herpesvirus 68 replication by selected antiviral agents. Antimicrob Agents Chemother 1998;42:170–172.
Hashemolhosseini S, Nagamine Y, Morley SJ et al. Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J Biol Chem 1998;273:14424–14429.
Buschhausen L, Kamm M, Arns W et al. Succssful treatment of a severe case of idiopathic pulmonary fibrosis with Rapamycin. Med Klinik 2005;100:161–164.
Korfhagen TR, Cras TDL, Davidson CR et al. Rapamycin prevents transforming growth factor-alpha-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2009;41:562–572.
Raghu G, Collard HR, Egan JJ et al. American Thoracic Society Documents. An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 2011;183:788–824.
Acknowledgements
We appreciate the assistance of Dr Margaret Nuovo, who did the photomicroscopy computer imaging. We also wish to thank the OSU Innovation Group for the Study of Complexity in Human, Natural, and Engineered Systems for covering the cost of publication. The work was supported by the Lewis Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Folcik, V., Garofalo, M., Coleman, J. et al. Idiopathic pulmonary fibrosis is strongly associated with productive infection by herpesvirus saimiri. Mod Pathol 27, 851–862 (2014). https://doi.org/10.1038/modpathol.2013.198
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.198
Keywords
This article is cited by
-
Human pluripotent stem cell-derived macrophages and macrophage-derived exosomes: therapeutic potential in pulmonary fibrosis
Stem Cell Research & Therapy (2022)
-
Emerging cellular and molecular determinants of idiopathic pulmonary fibrosis
Cellular and Molecular Life Sciences (2021)
-
Assessment of viral RNA in idiopathic pulmonary fibrosis using RNA-seq
BMC Pulmonary Medicine (2020)
-
Role of microbiota on lung homeostasis and diseases
Science China Life Sciences (2017)