Abstract
Spindle cell melanoma and desmoplastic melanoma differ clinically in prognosis and therapeutic implications; however, because of partially overlapping histopathological features, diagnostic distinction of spindle cell from desmoplastic melanoma is not always straightforward. A direct comparison of diagnostic and therapeutic biomarkers has not been performed. Meta-review of the literature discloses key clinicopathological differences between spindle cell and desmoplastic melanoma, including immunophenotypes. Using 50 biomarkers available in routine diagnostics, we examined 38 archival cases (n=16 spindle, 18 desmoplastic, 4 mixed spindle/desmoplastic melanoma). S100 remains as the most reliable routine marker to reach the diagnosis of melanoma in spindle cell and desmoplastic melanoma. We identified nine distinctly labeling markers with spindle cell melanoma showing positivity for laminin, p75, HMB45, c-kit, and MelanA, and desmoplastic melanoma preferentially labeling with collagen IV, trichrome, CD68, and MDM2. On the basis of comparisons of test performance measures, MelanA and trichrome were used to devise a 94% sensitive diagnostic algorithm for the distinction of desmoplastic from spindle cell melanoma. Gene amplification and expression status was assessed for a set of potentially drugable targets (HER2, EGFR, MET, MDM2, TP53, ALK, MYC, FLI-1, and KIT). Fluorescent in situ hybridizations did not reveal a significant number of gene aberrations/rearrangements; however, protein overexpression for at least one of these markers was identified in 35 of 38 cases (92%). In addition, we found BRAF mutations in 31% of spindle cell and 5% of desmoplastic melanoma, with an overall mutation frequency of 16% (n=6/38). We present the first comprehensive screening study of diagnostic and therapeutic biomarkers in spindle cell and desmoplastic melanoma. The devised algorithm allows diagnostic distinction of desmoplastic from spindle cell melanoma when routine histology is not decisive.
Similar content being viewed by others
Main
Spindle cell and desmoplastic melanoma are two distinct subtypes of malignant melanoma that differ by clinical, histopathological, and prognostic features.1 Histologically, both spindle cell and desmoplastic melanoma are characterized by atypical, spindled, malignant melanocytes. In desmoplastic melanoma, these malignant cells are intimately admixed with ropy and dense collagen fibrils. Anatomically, desmoplastic melanoma usually occurs in the head and neck region and presents clinically with a flat or nodular, scar-like lesion. Desmoplastic melanoma is usually not well circumscribed and malignant cells track along pre-existing structures such as skin appendages or nerves (eg, neurotropic melanoma). This specific growth pattern of desmoplastic melanoma is likely responsible for the higher rate of local recurrences, when compared with other melanoma subtypes.2, 3, 4, 5 There is an ongoing debate whether sentinel lymph node biopsy should be performed in desmoplastic melanoma because its metastatic rate is somewhat lower when compared with conventional melanoma and spindle cell melanoma.6, 7, 8, 9, 10, 11, 12 The overall prognosis in desmoplastic melanoma tends to be better when compared stage by stage with conventional melanoma.9, 13 In contrast, spindle cell melanoma is frequently amelanotic, can occur essentially anywhere on the body, and presents typically with widespread metastatic disease. Histologically, spindle cell melanoma is often confused with other (eg, mesenchymal or neural) tumors. This is why knowledge of specific immunophenotypes in spindle cell and desmoplastic melanoma can be helpful.9, 13, 14
Because of these differences, diagnostic distinction of spindle cell and desmoplastic melanoma is clinically relevant; however, currently mainly rests on the amount of scar-like tissue (ie, collagen content).1 Diagnostic distinction of spindle cell from desmoplastic melanoma is not always straightforward because of partially overlapping features and heterogeneity in collagen content. One approach to resolve this diagnostic challenge has been the introduction of an intermediate category (termed ‘mixed’ or ‘combined’ spindle/desmoplastic melanoma) that suggests a seemingly continuous biological spectrum.3, 10, 15, 16, 17, 18 Formally, the diagnostic categories in these overtly spindled melanomas are: spindle cell melanoma when the collagen content is <10%, mixed spindle/desmoplastic melanoma when collagen content is 10–90%, and desmoplastic melanoma when the collagen content is >90%.1, 16, 19, 20 From a practical perspective these formal definitions are problematic. First, superficial or partial biopsies may preclude assessment of the entire lesion. Second, lesional variability of the collagen content makes the assignment of one quantitative value difficult. Third, the immunoprofiles of spindle cell and desmoplastic melanoma differ not only from conventional melanoma but also between spindle cell and desmoplastic melanoma.20 These immunophenotypic differences indicate that collagen amount is not the only difference between desmoplastic and spindle cell melanoma. Collectively, this situation argues for a simple, reliable, and practical assay for diagnostic distinction of desmoplastic from spindle cell melanoma. It is surprising that despite the substantial body of work on immunophenotypes in spindle cell and desmoplastic melanoma, a direct comparative study of multiple diagnostic biomarkers has not been performed.
Here, we assessed a comprehensive biomarker panel for the distinction of desmoplastic from spindle cell melanoma and included biomarkers with potential therapeutic implications.
Materials and methods
Case Selection
After regulatory approval by the local Human Studies Committee, computer-based free-text queries were combined with ICD-based searches to identify patients with the diagnosis of spindle cell, mixed, or desmoplastic melanoma. After review and confirmation of the primary diagnosis according to established criteria1, 20 (H&E-based Ulm: SEW, BK, JKL; New York: BAH, DNS), cases with sufficient material were included. To minimize false diagnostic classification because of tumor heterogeneity, we selected primary excision specimen (35 of 38 cases) rather than biopsies or re-excisions whenever possible. The three included core-punch biopsies were also primary diagnoses and demonstrated classical features of spindle cell and desmoplastic melanoma (3 of 38 cases). We chose stringent selection criteria because morphology-based diagnoses served as the benchmark (‘gold standard’) for subsequent biomarker assessments. All samples had been formalin-fixed and paraffin-embedded. Masson trichrome staining was scored as positive when >10% of the lesional area consisted of blue collagen fibrils. The New York samples were freshly cut and unstained sections that were combined into arrays using a slide-to-slide transfer technique21 as follows. After deparaffinization, tissues were epoxy-resin-covered, lifted, microdissected into >2 mm2 tiles, and arrayed onto separate recipient slides. The staining performance using STS arrays and regular histological sections was confirmed in the one cohort (Ulm) before sections of the other cohort (New York) were arrayed, stained, and analyzed. For the Ulm cases, we used combinations of large-core (9 mm diameter) tissue microarrays, traditional 2-μm sections as well as slide-to-slide transfer arrays, capturing the superficial aspect including the epidermis whenever possible. Each array included control tissues composed of epithelial, neuronal, basement membrane, tonsillar, lymphatic, and collageneous tissue as well as conventional melanoma.
Immunohistochemistry
Immunohistochemistry was performed according to established protocols and details are provided in Supplementary Table 1. We ascertained specific staining using built-in controls (see above), negative controls (secondary antibody used without the primary antibody), and comparison of staining with prior reports and data from publicly available resources (http://www.proteinatlas.org; last accessed on 18 April 2013). All images were captured using an Olympus BX51 microscope connected to a digital camera or a whole-slide scanning system (.slide, both Olympus, Hamburg, Germany). Images were captured at 1440 × 900 pixel and cropped using Photoshop CS3 (Adobe Systems, San Jose, CA, USA).
Molecular Genetic Analysis
Fluorescent in situ hybridization (FISH) was performed to identify gene copy-number changes on formalin-fixed paraffin-embedded tissue by using a total of seven separate hybridizations. Probe details are provided in Supplementary Table 2. Hybridizations followed routine protocols and samples were counted and photographed using an Olympus BX60 Fluorescent Microscope (Olympus, Melville, NY, USA) with appropriate filter settings and software packages (MetaSystems, Altlussheim, Germany). Cutoffs for amplification probes followed definitions established for HER2 testing (gene copy-number ratio >2 scored in at least 50 nonoverlapping nuclei). Rearrangement was defined as splitting of otherwise fused yellow signals into separate green and red signals whereby the distance was apart >1 signal width. More than 15% of nuclei had to have split signals for a case to be scored as positive for a rearrangement. The 15% cutoff was based on counts of non-neoplastic controls, and represents the mean±3 s.d. of nuclei with split signals per 100 nuclei in normal tissue. For BRAF V600 genotyping, tumor regions were either sectioned and microdissected or cored using a 2-mm dermal punch needle. After deparaffinization, DNA extraction and PCR reactions (primers: R-5′-TGC TTG CTC TGA TAG GAA AAT G-3′; R-5′-AGC ATC TCA GGG CCA AAA AT-3′), pyrosequencing (R-5′-GAC CCA CTC CAT CGA G-3′) were performed on a PyroMarkQ24 sequencer (Qiagen, Hilden, Germany).22
Literature Review and Biomarker Selection
Two of the authors (SEW, JKL) performed a meta-review of the literature. Specific questions were: anatomic location by histotype, age at diagnosis, sex, and biomarker expression status. Both researchers reviewed a total of 193 published articles and independently extracted the predetermined criteria. After resolving differences via discussions, final tabulations were composed (see Supplementary Tables 5–9) and formed the starting point for our biomarker screening. We also included a set of therapeutically relevant markers from other tumor settings.
Statistical Analysis and Data Processing
For contingency testing, we applied Fisher’s exact test (association of histotype with dichotomous factors), χ2-test (association of histotype with >2 categories), or Student’s t-test (comparison of means). Data analysis was performed with Microsoft Excel 2008 (version 12.1.9; Microsoft, Redmond, WA, USA) or Prism 5.0b (GraphPad Softare, San Diego, CA, USA). Heatmaps were generated under the R programming language environment (http://www.r-project.org; version 2.13.2) and the pheatmap library. To sort all cases according to their overall biomarker phenotype, we performed unsupervised clustering analysis using the hclust function. On the basis of the maximum fraction of positive cases, each biomarker was assigned to one of two categories: ‘positive in spindle cell melanoma’ or ‘positive in desmoplastic melanoma’ (referred to as labeling direction). Differentially expressed biomarkers were defined as markers that showed significant differences in at least one cross validation. Cross validations compared differences within each cohort (eg, spindle cell melanoma-Ulm vs desmoplastic melanoma-Ulm), between cohorts (eg, spindle cell melanoma-Ulm vs spindle cell melanoma-New York), and/or by histotype (spindle cell all vs desmoplastic melanoma all). To estimate the s.d. of sensitivity and specificity for all differentially expressed biomarkers, we performed a leave-one-out analysis. The final decision tree for biomarker-based classification of spindle cell vs desmoplastic melanoma followed prior approaches23 and was based on considering the top three markers ‘positive in spindle cell melanoma’ and ‘positive in desmoplastic melanoma’, selecting for the highest sensitivity. Statistical performance measures were determined using the Hutchon toolkit (http://www.hutchon.net/EPRval.htm) and two-way contingency table analysis (http://www.statpages.org, last accessed on 18 April 2013). Statistical significance was defined as P<0.05.
Results
Archival searches, performed at two institutions, identified 38 cases composed of 16 spindle cell, 18 desmoplastic melanoma, and 4 mixed spindle/desmoplastic melanoma. Key clinicopathological parameters of each case are provided in Supplementary Table 3 and we combined the cohorts for contingency testing (Table 1) because characteristics did not differ between cohorts (Supplementary Table 4). In our series, desmoplastic melanoma was non-pigmented and more common in the head and neck region. To ensure the representativeness of our cohort, we performed a meta-review of 86 publications containing a total of 5955 spindle cell and desmoplastic melanoma cases (Supplementary Table 5). A summary along with our cases, including the anatomic distribution, is provided in Figure 1. Briefly, we found a male predominance (m-f-ratio: here 1.9:1 vs literature 1.8:1; P-range: 0.18–1.0) and the highest ratio of head and neck tumors in desmoplastic melanoma (here 52.6% vs literature 53.5%; P=1.0). Thus, by histopathological, epidemiologic, and clinicopatholgical parameters, we consider our cohort representative.
Synopsis of spindle, mixed, and desmoplastic melanoma. Anatomic distribution of current cases is provided along with the density calculated from a meta-review of the literature (see Supplementary Tables 5–9). Key features are provided below a representative microscopic field (H&E). Data taken from literature are provided in gray; the overall fraction of cases per diagnostic category is provided in brackets.
For biomarker screening, we performed a meta-review of 94 publications containing a total of 10 340 data points describing the reported immunophenotypes in spindle cell vs desmoplastic melanoma vs conventional melanoma (Supplementary Tables 6–8, respectively). Analysis of the positive fraction for each marker (Supplementary Table 9) revealed that desmoplastic melanoma, while being essentially negative for all classic melanocytic markers (eg HMB45, MelanA, and Tyrosinase), is S100 positive (96.4%), and expresses SOX10 (92.2%) and p75 (75%). These markers also label a significant fraction of spindle cell melanoma (S100 in 91.3%, SOX10 in 100%, and p75 in 79.5%) and can therefore be employed in reaching or confirming the diagnosis of melanoma. These literature data underscore a lack of markers that specifically label desmoplastic melanoma, which precludes reliable positive diagnostic distinction of desmoplastic from spindle cell melanoma (Figure 1). A limitation of this literature-based approach is that the percentage of positive cases is based on a theoretical summary of separate studies rather than a direct comparison of multiple markers in one study. The results of our direct comparative study of 50 routinely available biomarkers (38 IHC features, 10 FISH probes, one trichrome stain, and the BRAF V600 status) are provided in Figure 2a.
Biomarker screening of desmoplastic and spindle cell melanoma. (a) Results by marker (row) and histotype (columns); biomarkers with potential therapeutic relevance are indicated in red. Black box indicates biomarkers that showed distinct labeling in spindle cell vs desmoplastic melanoma (see Supplementary Tables 10 and 11). (b) Sensitivity and specificity plot for 10 diagnostically distinct biomarkers. Labels include the histotype that is positive for each maker; s.d. is derived from a leave-one-out analysis. C, cytoplasmic; N, nuclear; NT, non-tumor; T, tumor; V, vessels; (F), FISH; for details see Supplementary Tables 1 and 2.
We made two key findings when examining how spindle cell, desmoplastic, and mixed cases relate to the labeling pattern in a cluster analysis (Figure 3). First, three cases (one spindle cell, two desmoplastic) had a signature that was more similar to the opposite diagnostic category; however, re-review of the histopathology in these ‘outliers’ did not confirm the signature-based classification. Thus, we abstained from reclassification but included these ‘outliers’ in further analysis to take the apparent variation in immunophenotypes into account. Second, we found that the four mixed spindle/desmoplastic cases did not cluster together and, at least by signature-based similarity, had some phenotypic variability that covers the entire spindle cell-to-desmoplastic melanoma spectrum. To derive an algorithm for diagnostic distinction of desmoplastic from spindle cell melanoma, we argued that analysis of the marker pattern in the two extremes (spindle cell and desmoplastic melanoma only) would identify the most accurate markers and instead of reassignment of the mixed cases to spindle cell or desmoplastic categories, we excluded the four mixed cases from further analysis.
Case-based cluster analysis. Taking the entire biomarker phenotype into account, the resulting vertical order reflects similarity by phenotype and allows comparison with histotype. The position of the four mixed type melanomas is indicated along with three ‘outliers’ (arrows; see Results). C, cytoplasmic; D, desmoplastic melanoma; F, FISH/fluorescent in situ hybridization; M, mixed spindle and desmoplastic melanoma; .N, New York; N, nuclear; NT, non-tumor; S, spindle cell melanoma; T, tumor; .U, Ulm; V, vessels.
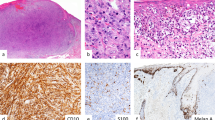
Systematic marker comparison in the 34 spindle cell and desmoplastic melanoma cases showed significant differences in 10 markers (Figure 2a, box and Supplementary Tables 10 and 11). Representative images comparing labeling patterns in spindle cell and desmoplastic melanoma are provided in Figure 4. Notably, the labeling direction of each marker has to be taken into account. Specifically, laminin (tumor and non-tumor), p75, HMB45, c-kit, and MelanA were significantly more often positive in spindle cell melanoma, whereas collagen IV, CD68, MDM2, and trichrome were significantly more often positive in desmoplastic than in spindle cell melanoma (Figure 2a). Briefly, collagen IV showed strong immunolabeling around individual tumor cells (Figure 4d), trichrome labeled the interspersed collagen fibrils (Figure 4h), CD68 demonstrated immunoreactivity in tumor cells (Figure 4l), and MDM2 showed nuclear labeling (Figure 4p). On the basis of test performance comparisons (Figure 2b), we chose the two markers with the highest sensitivity in spindle cell (ie, MelanA) and desmoplastic melanoma (ie, Trichrome) for the design of a diagnostic decision tree (see Discussion). Assessment in both cohorts revealed robust diagnostic distinction (Supplementary Table 11), and the overall test characteristics are shown in Table 2.
Diagnostically distinct biomarkers in spindle cell and desmoplastic melanoma. The figure is composed of two panels: six markers are positive in spindle cell melanoma and four markers are positive in desmoplastic melanoma. For each of the 10 biomarkers, we show one representative field in spindle cell (left: a, c, e, g, i, k, m, o, q, s) and desmoplastic melanoma (right: b, d, f, h, j, l, n, p, r, t); all images are taken at × 400. Abbreviations: NT, non-tumor; T, tumor.
As a part of the biomarker screen, we included a set of therapeutically relevant molecules (Figure 2a; red capsules). We assessed 10 markers with potential therapeutic relevance, and in case of HER2, EGFR, MET, MDM2, TP53, ALK, and MYC, we additionally performed FISH. We performed 266 hybridizations; however, only a single case with MDM2 amplification was identified (Figure 5a). Thus, from a practical standpoint, FISH-based genotyping for the included markers in spindle cell and desmoplastic melanoma has no value. We also assessed these potential targets by immunolabeling, and representative examples are provided in Figure 5. By fraction of positive cases, KIT and MYC were most frequently expressed in spindle cell melanoma, whereas TP53 and MDM2 were the most prevalent markers in desmoplastic melanoma. Although positive labeling for each marker was individually rare, the distribution of positive cases showed that several markers were expressed in a mutual exclusive fashion (eg, EGFR, MET, and HER2; Figure 2a), which adds up to a significant number of immunopositive cases (Table 3). Comparison of marker positivity in spindle cell and desmoplastic melanoma revealed that only KIT was significantly more commonly expressed in spindle cell melanoma. Although these findings are in line with an overall dearth of positive markers in desmoplastic melanoma, there were only three cases (two desmoplastic and one mixed) that were completely negative for any of the 10 potentially targetable molecules. Therefore, a total of 35 of 38 tumors in this cohort (92%) were positive for at least one biomarker with therapeutic significance in other tumor settings. Obviously, the practical significance of this finding requires validation; however, in conjunction with our FISH-based genotyping, it is safe to assume that the (ectopic) expression of these targetable molecules (Figures 5e, g, i and k) is the result of molecular mechanisms other than gene amplification (Figures 5d, f, h and j). Finally, we included BRAF genotyping (Figure 5c) and found V600 mutations in 31% of spindle cell and 5% of desmoplastic melanoma (Figure 2a). These numbers surmount to an overall mutation frequency of 16% (n=6/38) in spindle cell and desmoplastic melanoma, which, given the treatment implications in melanoma, argues for BRAF testing in this setting, when clinically indicated.
Selected biomarkers with potential therapeutic relevance in spindle cell and desmoplastic melanoma. Each marker (row) is shown as an image pair of fluorescence in situ hybridization (FISH; a, d, f, h, j); gene-to-copy-number ratio is provided on the bottom left) and immunohistochemistry (b, e, g, i, k), when applicable. (c) Pyrogram of case D-U1 shows ∼80% mutated BRAF alleles (as typically seen with loss of heterozygosity).
Discussion
Here, we present results of a comprehensive screening study of diagnostic and therapeutic biomarkers in spindle cell and desmoplastic melanoma. On the basis of the results in two independent cohorts, we devised an algorithm that allows diagnostic distinction of desmoplastic from spindle cell melanoma when histological analysis is not decisive (Figure 6).
In diagnostic practice, spindle cell and desmoplastic melanoma may reveal their identity as melanoma only after a first set of adjunct tests, typically including immunophenotyping. Importantly, spindle cell and desmoplastic melanoma may feature an atypical immunophenotype and even be negative for classic melanocytic markers. Thus, when confronted with a spindled lesion, demonstrating an atypical immunophenotype, the differential diagnosis should include melanocytic lesions and in particular spindle cell and desmoplastic melanoma. Our meta-review delineated that S100 and the less widely established markers SOX10 and p75 have the highest fraction of positivity in both spindle cell and desmoplastic melanoma (Supplementary Table 9). Thus, S100 remains as a reliable routine marker to reach the diagnosis of melanoma in spindle cell and desmoplastic melanoma.14 The prognostic differences in spindle cell and desmoplastic melanoma make their diagnostic distinction relevant; however, when lesions are partially sampled or heterogeneous, diagnosis may become difficult. In this setting—namely when a spindled lesion has been confirmed as a melanoma—our algorithm can be applied as an adjunct diagnostic tool. The diagnostic dilemma of spindle cell and desmoplastic melanoma14 extends, however, beyond atypical morphological and immunohistochemical features, and at least four additional points require consideration.
The Terminology Remains Confusing
Desmoplastic melanoma is currently defined as a subtype of spindle cell melanoma;1 however, the term spindle cell melanoma can either serve as an umbrella term for the spindle cell and desmoplastic melanoma category, might be applied as a descriptive diagnosis for a conventional melanoma with prominent spindle cells, or—as applied here—may refer to a specific entity that is typically amelanotic, contains <10% collagen fibrils, shows immunophenotypic differences to conventional melanoma,18, 24, 25, 26 and typically presents as a thicker lesion or in advanced stages. Further, BRAF mutations occur with different prevalence in spindle cell melanoma.27 These findings collectively suggest that the terminological imprecision of spindle cell melanoma may explain the variable incidence of 1–14% reported in the literature.28
The Prevalence of Spindle Cell and Desmoplastic Melanoma Maybe Higher Than Previously Assumed
Our meta-review from the first description by Conley29 in 1971 until April 2013 lists nearly 6000 cases, arguing that spindle cell and desmoplastic melanoma cannot be considered orphan diseases anymore.
The Diagnostic Categories are Part of an Extended Biological Spectrum
The partly overlapping morphological features along with clinicopathological differences have led to the introduction of a mixed spindle and desmoplastic melanoma category.3, 15, 16, 17 Briefly, mixed spindle cell and desmoplastic melanoma is defined as composed of spindled melanocytes admixed with 10–90% collagen fibrils. Although the distinct immunoprofile for Ki-67, CD117, and Nestin in mixed spindle cell and desmoplastic melanoma has been reported,30 we abstained from inclusion of mixed cases into our analysis because of the small number (n=4 in our cohorts), the phenotypic variability (see Results), and to establish the decision tree based on findings in the extremes (ie, ‘pure’ spindle cell vs desmoplastic melanoma). This should, however, not imply that the clinically distinct mixed subtype ought to be neglected.3, 15, 16, 17 Rather we emphasize that our algorithm is not a one size-fits-all approach but allows inclusion of existing or additional categories. The concept of an extended spectrum is also supported by comparative genome hybridization data demonstrating that 67% of desmoplastic melanoma share allelic losses of NF1, which matches the immunophenotypic overlap of desmoplastic melanoma with malignant peripheral nerve sheet tumors.26, 31, 32, 33, 34 Collectively, these data suggest that spindle cell and desmoplastic melanoma represent discrete diagnostic categories within an extended biological spectrum.
There is a Dearth of Specific Desmoplastic Melanoma Markers
Gene expression profiling in desmoplastic melanoma revealed at least 629 differentially expressed genes35 that have been (in part) validated by immunohistochemistry. However, the overall rarity of desmoplastic melanoma may not justify implementation of novel antibodies in every laboratory. Thus, we focused on routinely available and established markers and extend the list of specific desmoplastic melanoma markers. We identified MDM2 as positive in desmoplastic melanoma (albeit with less optimal test performance characteristics) and a subset of desmoplastic melanoma that is CD68 positive. The latter is important because one key differential diagnosis of desmoplastic melanoma is a scar that typically contains CD68-positive histiocytes.36 In addition, CD68 may not be helpful in differentiating histiocytic tumors from desmoplastic melanoma.37 Whether CD68 can become important in terms of classification remains to be determined. Currently, the reason for the ectopic CD68 expression remains unclear20, 38, 39 and we interpret this histiocytic feature of desmoplastic melanoma as part of the ‘great imitator’ malignant melanoma. We also identified collagen IV around tumor cells as a relatively sensitive and specific phenomenon in desmoplastic melanoma (Figure 4d). It seems unlikely that this staining pattern has not been previously noted in desmoplastic melanoma;33 however, peritumoral collagen IV labeling is in accord with the overall histomorphology and indicates that each tumor cells synthesized at least one principle component of the surrounding basement membrane, essentially providing its own structural encasement. As the addition of collagen IV did not increase test performance when trichrome staining was included, we chose the special stain as a cost-effective and reliable approach. Thus, two readily available markers (MelanA and trichrome staining) achieved the best diagnostic performance; however, the presented algorithm should not replace careful histomorphological examination and clinicopathological correlation.
In summary, this is the first literature-based, comprehensive screening study that compares the profile of diagnostic and potentially targetable molecules in spindle cell vs desmoplastic melanoma with respect to commonly used tissue biomarkers. A panel approach identifies a significant fraction of patients that may benefit from molecularly targeted therapy and the presented diagnostic algorithm may guide classification when histopathological classification is not straightforward.
References
McCarthy SW, Crotty KA, Scolyer RA . Desmoplastic melanoma and desmoplastic neurotropic melanoma In: LeBoit PE (eds) Pathology and genetics of skin tumours. IARC Press: Lyon, France, 2006.
Chen JY, Hruby G, Scolyer RA et al. Desmoplastic neurotropic melanoma: a clinicopathologic analysis of 128 cases. Cancer 2008;113:2770–2778.
Hawkins WG, Busam KJ, Ben-Porat L et al. Desmoplastic melanoma: a pathologically and clinically distinct form of cutaneous melanoma. Ann Surg Oncol 2005;12:207–213.
Jaroszewski DE, Pockaj BA, DiCaudo DJ et al. The clinical behavior of desmoplastic melanoma. Am J Surg 2001;182:590–595.
Maurichi A, Miceli R, Camerini T et al. Pure desmoplastic melanoma: a melanoma with distinctive clinical behavior. Ann Surg 2010;252:1052–1057.
Broer PN, Walker ME, Goldberg C et al. Desmoplastic melanoma: a 12-year experience with sentinel lymph node biopsy. Eur J Surg Oncol 2013;39:681–685.
Gyorki DE, Busam K, Panageas K et al. Sentinel lymph node biopsy for patients with cutaneous desmoplastic melanoma. Ann Surg Oncol 2003;10:403–407.
Lens MB, Newton-Bishop JA, Boon AP . Desmoplastic malignant melanoma: a systematic review. Br J Dermatol 2005;152:673–678.
Livestro DP, Muzikansky A, Kaine EM et al. Biology of desmoplastic melanoma: a case-control comparison with other melanomas. J Clin Oncol 2005;23:6739–6746.
Mohebati A, Ganly I, Busam KJ et al. The role of sentinel lymph node biopsy in the management of head and neck desmoplastic melanoma. Ann Surg Oncol 2012;19:4307–4313.
Murali R, Shaw HM, Lai K et al. Prognostic factors in cutaneous desmoplastic melanoma: a study of 252 patients. Cancer 2010;116:4130–4138.
Shaw HM, Quinn MJ, Scolyer RA et al. Survival in patients with desmoplastic melanoma. J Clin Oncol 2006;24:e12 author reply e3.
de Almeida LS, Requena L, Rutten A et al. Desmoplastic malignant melanoma: a clinicopathologic analysis of 113 cases. Am J Dermatopathol 2008;30:207–215.
Ohsie SJ, Sarantopoulos GP, Cochran AJ et al. Immunohistochemical characteristics of melanoma. J Cutan Pathol 2008;35:433–444.
Busam KJ, Mujumdar U, Hummer AJ et al. Cutaneous desmoplastic melanoma: reappraisal of morphologic heterogeneity and prognostic factors. Am J Surg Pathol 2004;28:1518–1525.
McCarthy SW, Scolyer RA, Palmer AA . Desmoplastic melanoma: a diagnostic trap for the unwary. Pathology 2004;36:445–451.
Scolyer RA, Thompson JF . Desmoplastic melanoma: a heterogeneous entity in which subclassification as "pure" or "mixed" may have important prognostic significance. Ann Surg Oncol 2005;12:197–199.
Skelton HG, Maceira J, Smith KJ et al. HMB45 negative spindle cell malignant melanoma. Am J Dermatopathol 1997;19:580–584.
Coupelon S, Franck F, Jarrousse AS et al. Desmoplastic malignant melanoma: a study of ten cases and status of BRAF mutation. Dermatology 2012;225:168–171.
Longacre TA, Egbert BM, Rouse RV . Desmoplastic and spindle-cell malignant melanoma. An immunohistochemical study. Am J Surg Pathol 1996;20:1489–1500.
Weissinger SE, Moeller P, Lennerz JK . Slide-to-slide arrays for high-throughput molecular profiling or rare tumor specimens. Mod Pathol 2013;26:503A.
Lennerz JK, Klaus BM, Marienfeld RB et al. Pyrosequencing of BRAF V600E in routine samples of hairy cell leukaemia identifies CD5+ variant hairy cell leukaemia that lacks V600E. Br J Haematol 2012;157:267–269.
Choi WW, Weisenburger DD, Greiner TC et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res 2009;15:5494–5502.
Mohamed A, Gonzalez RS, Lawson D et al. SOX10 expression in malignant melanoma, carcinoma, and normal tissues. Appl Immunohistochem Mol Morphol 2013, (in press).
Pages C, Rochaix P, al Saati T et al. KBA.62: a useful marker for primary and metastatic melanomas. Hum Pathol 2008;39:1136–1142.
Xu X, Chu AY, Pasha TL et al. Immunoprofile of MITF, tyrosinase, melan-A, and MAGE-1 in HMB45-negative melanomas. Am J Surg Pathol 2002;26:82–87.
Kim J, Lazar AJ, Davies MA et al. BRAF, NRAS and KIT sequencing analysis of spindle cell melanoma. J Cutan Pathol 2012;39:821–825.
Piao Y, Guo M, Gong Y . Diagnostic challenges of metastatic spindle cell melanoma on fine-needle aspiration specimens. Cancer 2008;114:94–101.
Conley J, Lattes R, Orr W . Desmoplastic malignant melanoma (a rare variant of spindle cell melanoma). Cancer 1971;28:914–936.
Miller DD, Emley A, Yang S et al. Mixed versus pure variants of desmoplastic melanoma: a genetic and immunohistochemical appraisal. Mod Pathol 2011;25:505–515.
Boyle JL, Haupt HM, Stern JB et al. Tyrosinase expression in malignant melanoma, desmoplastic melanoma, and peripheral nerve tumors. Arch Pathol Lab Med 2002;126:816–822.
Kanik AB, Yaar M, Bhawan J . p75 nerve growth factor receptor staining helps identify desmoplastic and neurotropic melanoma. J Cutan Pathol 1996;23:205–210.
Prieto VG, Woodruff JM . Expression of basement membrane antigens in spindle cell melanoma. J Cutan Pathol 1998;25:297–300.
Pryor JG, Brown-Kipphut BA, Iqbal A et al. Microarray comparative genomic hybridization detection of copy number changes in desmoplastic melanoma and malignant peripheral nerve sheath tumor. Am J Dermatopathol 2011;33:780–785.
Busam KJ, Zhao H, Coit DG et al. Distinction of desmoplastic melanoma from non-desmoplastic melanoma by gene expression profiling. J Invest Dermatol 2005;124:412–418.
Ramos-Herberth FI, Karamchandani J, Kim J et al. SOX10 immunostaining distinguishes desmoplastic melanoma from excision scar. J Cutan Pathol 2010;37:944–952.
Pernick NL, DaSilva M, Gangi MD et al. "Histiocytic markers" in melanoma. Mod Pathol 1999;12:1072–1077.
Morgan MB, Purohit C, Anglin TR . Immunohistochemical distinction of cutaneous spindle cell carcinoma. Am J Dermatopathol 2008;30:228–232.
Shah IA, Gani OS, Wheler L . Comparative immunoreactivity of CD-68 and HMB-45 in malignant melanoma, neural tumors and nevi. Pathol Res Pract 1997;193:497–502.
Acknowledgements
We thank Dr Ulrike Kostezka, Michaela Buck, Julia Kiedaisch, and Iwona Nerbas for expert technical assistance. We also thank Uwe Gerstenmaier (Varionostics, Ulm, Germany) for help with genotyping and Karin Löschner (Anatomy LS1, Erlangen, Germany) for sharing of an antibody during biomarker selection. JKL was supported by the Else-Kröner Fresenius Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Weissinger, S., Keil, P., Silvers, D. et al. A diagnostic algorithm to distinguish desmoplastic from spindle cell melanoma. Mod Pathol 27, 524–534 (2014). https://doi.org/10.1038/modpathol.2013.162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.162