Abstract
Well-differentiated small intestinal neuroendocrine tumors are rare malignancies. They arise from enterochromaffin cells and very little is known about differential microRNA (miRNA) expression. The aim of this study was to identify the miRNA profile of well-differentiated small intestinal neuroendocrine tumors, which may have a critical role in tumor development, progression and potentially develop miRNAs as novel clinical biomarkers. Specimens from two test groups, 24 small intestinal neuroendocrine tumor specimens at different stages of malignancy, are included in this study. Total RNA from the first test group, five primary tumors, five mesentery metastases and five liver metastases was hybridized onto the Affymetrix Genechip miRNA arrays to perform a genome-wide profile. The results were validated by using quantitative real-time PCR (QRT-PCR) and northern blot analyses. We then expanded the investigation to laser capture microdissected small intestinal neuroendocrine tumor cells and immuno-laser capture microdissected normal enterochromaffin cells of the first test group. Furthermore, a second test group, three primary tumors, three mesentery metastases and three liver metastases, was included in the study. Thus, two independent test groups validated the data by QRT-PCR. Moreover, we characterized nine miRNAs, five (miR-96, -182, -183, -196a and -200a), which are upregulated during tumor progression, whereas four (miR-31, -129-5p, -133a and -215) are downregulated. Several online software programs were used to predict potential miRNA target genes to map a number of putative target genes for the aberrantly regulated miRNAs, through an advanced and novel bioinformatics analysis. Our findings provide information about pivotal miRNAs, which may lead to further insights into tumorigenesis, progression mechanisms and novel therapeutic targets recognition.
Similar content being viewed by others
Main
Small intestinal neuroendocrine carcinomas arise from neuroendocrine cells. They are classified according to differentiation status and tumor cell type, and some of them are designated as well-differentiated small intestinal neuroendocrine tumors according to the WHO classification, 2010.1, 2 The human neuroendocrine cells are part of the diffuse endocrine cell system. They are either confined to certain organs, such as thyroid, pancreas or adrenals, or dispersed throughout the body mainly in the respiratory tract and in the intestinal mucosa. The cells can accumulate precursor molecules, which are stored and then processed into hormones or amines. The hormones and amines are released on stimulation either to the blood stream or to adjacent cells or neurons. Furthermore, in the intestinal tract mucosa they can be released into the intestinal lumen. However, the release is always tightly controlled, as peptides and amines regulate a variety of processes, such as gastrointestinal secretion, motility and blood pressure in the human body.3, 4
The largest group of neuroendocrine tumors is the gastroenteropancreatic neuroendocrine tumors followed by lung neuroendocrine tumors. The small intestinal neuroendocrine tumors belong to gastroenteropancreatic neuroendocrine tumors and arise from enterochromaffin cells, which are neuroendocrine cells disseminated throughout the gastrointestinal tract. The majority of the small intestinal neuroendocrine tumor patients get diagnosis when they have already developed metastatic malignancies and no curative treatment is currently available.5, 6 However, tumor biology has shown recent advancement in small intestinal neuroendocrine tumors’ immunotherapy and virotherapy7, 8, 9, 10 in combination with more recent findings to detect novel small intestinal neuroendocrine tumor biomarkers.11, 12
Over the past decade, a variety of gene regulatory mechanisms, which are controlled by a large class of non-coding gene, have been unveiled. They transcribe small RNAs known as microRNAs (miRNA), which are mostly transcribed from intragenic or intergenic regions. The primary transcript undergoes further processing by two pivotal ribonucleases in the nucleus, resulting in a hairpin intermediate of about 70–100 nucleotides (nt), called pre-miRNA, which is exported to the cytoplasm. The pre-miRNA is then processed and reduced into a mature double-stranded RNA of 18–25 nt by a different ribonuclease. The double-stranded RNA contains the guide strand and the passenger strand. After the strands separate, the guide strand will be incorporated into an RNA-induced silencing complex (RISC), whereas the passenger strand is degraded. The mature strand is pivotal for target recognition and incorporation of the specific target mRNA into the RISC.13
MiRNAs function as repressor of eukaryotic gene regulation by binding to the 3′-untranslated regions of target mRNAs. The binding triggers cleavage or translational inhibition of the mRNA, depending on the degree of complementarities.14, 15 Several hundred miRNAs are present in mammals and regulate the expression of many genes. Indeed, each miRNA is potentially able to target a large number of genes, which are involved in different cellular mechanisms, such as cell proliferation, apoptosis, differentiation, viral infection and tumorigenesis.16, 17
Many studies have previously revealed that malignant tissues in cancer patients exhibit distinctive miRNA expression signatures.18 Moreover, genome-wide profiling showed that these signatures discriminate different types of cancer with a high degree of accuracy. However, miRNA profiles can sometimes mislead cancer type distinction.13 Despite the advances in understanding the mechanisms, which cause miRNA aberrant regulation, the most challenging task is to elucidate the biological role of miRNAs in cancer initiation and development.17 A growing number of potential oncogenic and suppressor miRNAs have been identified, which support the use of specific miRNA signatures to predict clinical outcome of lung neuroendocrine tumors19, 20, 21 and more recently of small intestinal neuroendocrine tumors.22 Although the recent findings have been enhanced by an animal model result,23 very little has been elucidated about human small intestinal neuroendocrine tumors’ differential gene and miRNA expression.22
To address this challenge, we performed miRNA profiling with Affymetrix GeneChip miRNA arrays of small intestinal neuroendocrine tumors at different stages of disease. Furthermore, miRNA analysis was extended to laser capture microdissected normal enterochromaffin cells and tumor cells to improve the specific miRNA cellular expression data. Moreover, bioinformatics analysis and advanced molecular biology methods were used to identify specifically and differentially expressed miRNAs, which may be developed as novel biomarkers and therapeutic molecular targets.
Materials and methods
Tissue Samples
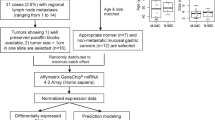
Two independent test groups of 24 snap-frozen specimens are denoted by numbers. The first group includes 15 specimens (1–15), whereas the second one includes 9 specimens (16–24). Tissue blocks are from patients, with a histopathologically confirmed diagnosis of small intestinal neuroendocrine tumors at different stages of disease (eight primary tumors, eight mesentery metastases and eight liver metastases). Patients’ information is summarized in Table 1. Permission to collect carcinoma specimens was approved by the regional Ethical Committee at the Uppsala University Hospital (Dnr 2011/426).
RNA Extraction
Total RNA was isolated from tissue specimens, by using mirVana miRNA Isolation Kit (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions, and total RNA from immuno-laser capture microdissected normal enterochromaffin cells is a generous gift by the RV Lloyd group, as described previously.12 RNAqueous Micro Kit (Life Technologies) was used to prepare total RNA from microdissected cells. RNA quality and quantity was verified by using the RNA 6000 Nano Kit/RNA 6000 Pico Kit and the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Agilent Small RNA Kit was used to verify miRNA quality and quantity by using Agilent 2100 Bioanalyzer (Agilent Technologies).
MicroRNA Arrays’ Data Analysis and Data Mining
About 1 μg of total RNA from small intestinal neuroendocrine tumor snap-frozen specimens described above was sent to the Bioinformatics and Expression Analysis (BEA) core facility (Karolinska Institute, Huddinge, Sweden). Total RNA was hybridized onto the Affymetrix GeneChip miRNA 1.0 Array (Affymetrix, Santa Clara, CA, USA) and processed according to Affymetrix technical protocols. Scanned images of microarray chips were analyzed by Affymetrix miRNA QC Tool (Affymetrix). The raw data have been normalized by using a quantile algorithm. Expression levels in the different tissues have been compared by using two-sided unpaired Student’s t-test, and false discovery rates (q-values) were estimated by q-value package in R (www.r-project.org). The hierarchical clustering was performed by using the MeV software (www.tm4.org).24 Differentially expressed miRNAs were identified by calculating fold change of either mesentery metastasis group versus primary carcinoma group or liver metastasis group versus primary tumor group. The hierarchical clustering algorithm has been applied to the group of miRNAs from the 15 tumor samples according to similarities in expression, which have been clustered according to Pearson’s correlation distance. The results from the miRNA array analysis have been deposited to the National Center for Biotechnology Information (NCBI)’s GEO (accession number: GSE33568) and the European Bioinformatics Institute (EBI)’s Array Express (accession number: E-MTAB-862).
QRT-PCR
In all, 1 μg of total RNA from small intestinal neuroendocrine tumor snap-frozen specimens and 1 ng of total RNA from laser capture microdissected cells were reverse transcribed and converted to cDNA by QuantiMir RT Kit (System Biosciences, Mount View, CA, USA) according to the manufacturer’s instructions. The cDNAs were used to analyze miRNA expression, by using primers described in Supplementary Table 1. MiR-31, -129-5p, -133a, -182, -183, -196a, -200a and -215 expression was measured by using Stratagene Mx3005P real-time PCR System (Agilent Technologies) and Brilliant SYBR Green QPCR Master Mix (Agilent Technologies). The 2−ΔΔCT method25 evaluated the data by using RNU48 (a small nucleolar RNA) set to 1 as endogenous control.
Northern Blot Analysis
Northern blot analysis was performed using mirVana miRNA Detection Kit (Applied Biosystems), which is hybridization-based solution to analyze miRNA expression, according to the manufacturer’s instructions. Total RNA (5 μg) from snap-frozen small intestinal neuroendocrine tumor specimens of primary tumors, mesentery metastases and liver metastases were incubated with a 32P-labeled RNA probe (Perkin-Elmer, Boston, MA, USA) followed by RNase digestion. The radioisotope-labeled RNA fragment were prepared by using mirVana miRNA Probe Construction Kit (Applied Biosystems) according to the manufacturer’s instructions. The following probes were used: miR-31 (5′-AGGCAAGATGCTGGCATAGCTCCTGTCTC-3′), miR-133a (5′-TTTGGTCCCCTTCAACCAGCTGCCTGTCTC-3′), miR-182 (5′-TTGGCAATGGTAGAACTCACACTCCTGTCTC-3′), miR-183 (5′-TATGGCACTGGTAGAATTCACTCCTGTCTC-3′) and RNU48 (5′-GATGACCCCAGGTAACTCTGAGTGTGTCGCTGATGCCATCACCGCAGCGCTCTGACCCCTGTCTC-3′). The radiolabeled RNA samples were analyzed on 15% TBE-Urea gels (Bio-Rad, Hercules, CA, USA) and detected by Fuji radiography film (Fujifilm, Tokyo, Japan). The developed radiography films of Northern blot analyses were scanned for the semiquantitative analysis using the Molecular Imager Chemidoc XRS+ System (Bio-Rad) and the Image Lab Software (Bio-Rad).
Immuno-Laser Capture Microdissection of Normal Enterochromaffin Cells
Laser capture microdissection was combined with immunocytochemical staining with specific antibodies to identify endocrine cells associated with the small intestinal crypts. Snap-frozen specimens from normal ileum mucosa were cut in 10 μm sections by a microtome cryostat (Leica, Wetzlar, Germany) and adhered on uncharged glass slides and cells with a positive immunostaining signal cells were isolated as described previously.12, 26 The slides were incubated with anti-human chromogranin A (CgA) antibody (1:100; Dako, Santa Barbara, CA, USA), washed and incubated with a secondary horseradish peroxidase-conjugated antibody, washed and counterstained with hematoxylin. All steps were performed at the minimum time required and in the presence of RNase inhibitor (Life Technologies). We have previously verified that the CgA antibody can be combined with laser capture microdissection, as shown by Leja et al.12 Enterochromaffin cells are located in the crypts within the ileum and we estimate that one endocrine cell and three to four non-endocrine cells were captured with each laser treatment, that is, approximately 25% of the captured cells are endocrine and the majority of them are enterochromaffin cells.12 Immunostained cells were isolated using a PixCell II LCM system (Arcturus Bioscience, Mount View, CA, USA).
Laser Capture Microdissection of Neuroendocrine Tumor Cells
Nine specimens (three primary tumors, three mesentery metastases and three liver metastases) from the first test group, and nine specimens (three primary tumors, three mesentery metastases and three liver metastases) from the second test group were used to perform laser capture microdissection of small intestinal neuroendocrine tumor cells. The snap-frozen specimens were cut in 10 μm sections by a microtome cryostat (Leica) and adhered to polyethylene-naphtalate membrane frame slides (Life Technologies). Stromal cells from primary tumor blocks and tumor cells from primary tumors, and mesentery and liver metastatic cells were isolated by ArcturusXT Microdissection System (Life Technologies) according to the manufacturer’s instructions.
Prediction of MicroRNA Targets
Several online software programs were used to investigate the predicted target genes, such as DIANA LAB (http://diana.cslab.ece.ntua.gr/), PicTar (http://pictar.mdc-berlin.de/), miRDB (http://mirdb.org/miRDB/) and TargetScan (http://www.targetscan.org/). Potential miRNA target genes were predicted by using the online software programs mentioned above in combination with the published small intestinal neuroendocrine tumor microarray profiling data published by our group.12 The main principles of miRNA target prediction are from the conserved sites that match the seed region of each miRNA.27 The biological functions of the potential miRNA targets were further predicted by using Ensembl (http://www.ensembl.org/) gene ontology information.
Statistical Analysis
All experiments were performed in triplicate. The statistical significance of the difference between two groups was evaluated either by one-way ANOVA followed by Bonferroni test or by two-tailed unpaired Student’s t-test using GraphPad Prism 5 (GraphPad Software, La Jolla CA, USA); P-value<0.05 is considered significant.
Results
Cluster Analysis Reveals 33 Differentially Expressed miRNAs in Small Intestinal Neuroendocrine Tumors at Different Stage of Disease
We profiled five primary tumors (P1–5), five mesentery metastases (M1–5) and five liver metastases (L1–5) by using Affymetrix miRNA arrays. We first interpreted the raw data and created a cluster dendrogram of miRNA expression profiles from small intestinal neuroendocrine tumor specimens at different stages of disease. We then filtered the genome-wide Affymetrix miRNA array analysis to restrict 33 differentially expressed miRNAs in mesentery metastases versus primary tumors. In addition, we filtered the same number of differentially expressed miRNAs in liver metastases versus primary tumors. Figure 1a shows that 15 miRNAs are upregulated, whereas 18 are downregulated. Moreover, the 33 miRNAs are differentially expressed in the two different metastatic stages. Furthermore, a specific absolute variance is clearly evident in the results of Figure 1a. Indeed, 14 out of 15 upregulated miRNAs increased more in the liver metastases than in the mesentery metastases compared to the primary tumors with the unique exclusion of miR-148a. Figure 1a shows the 15 upregulated miRNA (yellow bars), whereas the 18 downregulated miRNAs are shown by blue bars. Nine miRNAs, which have been chosen for further validation, are indicated by a red asterisk. Primary tumors cluster together differently from the metastatic ones. Thus, 9 out of 10 metastatic samples are distant from the primary tumors and cluster together, as shown by the blue triangle in Figure 1b. However, M2 is a unique exception (Figure 1b).
Representative 33 differentially expressed microRNAs (miRNAs) from frozen well-differentiated small intestinal neuroendocrine tumor specimens by using genome-wide Affymetrix miRNA arrays. (a) In all, 33 differentially expressed miRNAs were detected from mesentery metastases (M) and liver metastases (L) compared with primary tumors (P). Out of 33 miRNAs, 9 were selected for further analysis and they are marked by a red asterisk. Deregulated miRNAs expression is plotted in yellow (up) and in blue (down). (b) A cluster of 33 differentially expressed miRNAs from five P (P1–5), five M (M1–5) and five L (L1–5) is shown as an example. Out of 10 metastatic samples, 9 were clustered together (indicated by the blue triangle in the figure) with a singular exception of sample M2. MiRNAs were clustered according to Pearson’s correlation distance as indicated in the figure.
QRT-PCR and Northern Blot Analyses Confirmed miRNA Arrays In Silico Data
We further validated our data by using QRT-PCR and Northern blot analyses. We first investigated the nine selected miRNAs using frozen specimens from the first test group, which were profiled as described above, by using QRT-PCR analysis. Five miRNAs (miR-96, -182, -183, -196a and -200a) appear increased in expression by comparing primary tumors to both mesentery and liver metastases. However, only four out of five (miR-96, -182, -183 and -196a) are significantly upregulated, whereas four (miR-31, -129-5p, -133a and -215) are significantly downregulated (Figures 2a and b). In addition, we detected variation of the evaluated miRNAs in the small intestinal neuroendocrine tumors by using Northern blot analyses (Figure 3a) with quantification of expression ratios in Figure 3b. Indeed, miR-182 increases from the primary tumor stage to mesentery metastasis stage, peaking at the liver metastasis stage precisely following the tumor progression stages. MiR-183 expression increases at the liver metastasis stage and does not differ between the primary tumors and mesentery metastasis stages. MiR-31 and -133a expression is clearly downregulated both at the mesentery and liver metastasis stages compared with primary tumors.
Quantitative real-time PCR (QRT-PCR) analysis validated the expression of the nine selected microRNA (miRNAs) from the first group of specimens. Total RNA was isolated from frozen specimens of three primary tumors (P), three mesentery metastases (M) and three liver metastases (L) to run QRT-PCR analysis. (a) Upregulated miRNA expression in metastatic disease compared with primary tumors. (b) Downregulated miRNA expression in metastatic disease compared with primary tumors. Results were plotted using the 2−ΔΔCt method with RNU48 expression (set to 1) from each individual sample for normalization. Plotted results are mean±s.d. for triplicate wells. Significance was calculated by one-way analysis of variance (ANOVA) followed by Bonferroni test. *P<0.05, **P<0.01 and ***P<0.001.
Northern blot analysis validated miR-31, -133a, -182 and -183 expression. Total RNA was isolated from frozen well-differentiated small intestinal neuroendocrine tumor specimens, which were primary tumor (P), mesentery metastasis (M) and liver metastasis (L). (a) Specific 32P-labeled probes were hybridized to total RNA. (b) The fold changes were calculated as the ratio of mesentery metastasis versus primary tumor and liver metastasis versus primary tumor. RNU48 was used as an internal control to standardize the calculation of fold change.
QRT-PCR Analyses on Immuno-Microdissected Normal Enterochromaffin Cells and Microdissected Small Intestinal Neuroendocrine Tumor Cells Confirm Expression Variation of the Nine Selected miRNAs Between Normal Cells and Tumor Cells
We conducted QRT-PCR on total RNA, from immuno-microdissected normal enterochromaffin cells and microdissected small intestinal neuroendocrine tumor cells at different stage of disease, three primary tumors, three mesentery metastases and three liver metastases. Thus, we verified that five miRNAs (miR-96, -182, -183, -196a and -200a) are significantly upregulated (Figure 4a), whereas four miRNAs (miR-31, -129-5p, -133a and -215) are significantly downregulated (Figure 4b).
Quantitative real-time PCR (QRT-PCR) analysis validated the expression of nine selected microRNAs (miRNAs) from the first test group. Total RNA was isolated from microdissected tumor cells and microdissected normal enterochromaffin cells. Analysis was run using three normal enterochromaffin cell, three primary tumor, three mesentery metastasis and three liver metastasis samples. (a) Upregulated miRNA expression in tumor cells compared with normal enterochromaffin cells. (b) Downregulated miRNA expression in tumor cells compared with normal enterochromaffin cells. Results were plotted using the 2−ΔΔCt method with RNU48 expression (set to 1) from each individual sample for normalization. Plotted results are mean±s.d. for triplicate wells. Significance was calculated by one-way analysis of variance (ANOVA) followed by Bonferroni test. *P<0.05, **P<0.01 and ***P<0.001.
Briefly, miRNA expression of miR-96 is on average 100-fold higher in tumor cells at different stage than in normal enterochromaffin cells; miR-182 is expressed 1000-fold more in the tumor cells than in normal enterochromaffin cells. MiR-196a is on average 100-fold higher in primary tumor and mesentery metastatic cells than in normal enterochromaffin cells, whereas it is on an average 1000-fold higher in liver metastatic cells. MiR-183 and -200a show a higher basal expression in normal enterochromaffin cells than miR-96, -182 and -196a. However, the average expression level is still 10-fold higher in primary tumor and mesentery metastatic cells and 100-fold higher in liver metastatic cells than in normal enterochromaffin cells (Figure 4a). The results for the downregulated miRNAs show that the expression level of miR-31 is approximately 1000-fold lower in tumor cells than in normal enterochromaffin cells, which express high levels of this miRNA. MiR-129-5p shows a slight downregulation in tumor cells compared with normal enterochromaffin cells, on an average around 10-fold. MiR-133 is on an average 10-fold lower in primary tumor cells and 100-fold in metastatic cells than in normal enterochromaffin cells. MiR-215 shows a similar differential expression as miR-31 (Figure 4b).
QRT-PCR Analysis on the Second Test Group Confirmed the Results of the First Test Group Analysis on the Nine Selected miRNAs
We conducted a second independent QRT-PCR analysis on total RNA by using the second test group. Thus, we included immuno-microdissected normal enterochromaffin cells and microdissected small intestinal neuroendocrine tumor cells, from three primary tumors, three mesentery metastases and three liver metastases. The results are in agreement with that described previously and as also shown in Figures 4a and b. Indeed, miR-96, -182, -183, -196a and -200a are significantly upregulated in the second screening, which is shown in Figure 5a, whereas miR-31, -129-5p, -133a and -215 are significantly downregulated, which is shown in Figure 5b. The analogous results do not require any further explanation as they can be easily extrapolated looking at Figures 4a and b according to our opinion.
Quantitative real-time PCR (QRT-PCR) analysis validated the expression of nine selected microRNAs (miRNAs) from the second test group. Total RNA was isolated from microdissected tumor cells and microdissected normal enterochromaffin cells. Analysis was run using three normal enterochromaffin cell, three primary tumor, three mesentery metastasis and three liver metastasis samples. (a) Upregulated miRNA expression in tumor cells compared with normal enterochromaffin cells. (b) Downregulated miRNA expression in tumor cells compared with normal enterochromaffin cells. Results were plotted using the 2−ΔΔCt method with RNU48 expression (set to 1) from each individual sample for normalization. Plotted results are mean±s.d. for triplicate wells. Significance was calculated by one-way analysis of variance (ANOVA) followed by Bonferroni test. *P<0.05, **P<0.01 and ***P<0.001.
Expression Variation Significance of Nine miRNAs on Laser Capture Microdissected Stromal Cells Versus Laser Capture Microdissected Primary Tumor Cells
We analyzed whether the variation of miRNAs, expressed in microdissected stromal cells versus microdissected primary tumor cells, was statistically significant. This is a pivotal investigation as it is of major scientific importance in the evaluation whether the miRNAs are more highly expressed in microdissected enriched tumor cells than in the microdissected enriched adjacent stromal cells. The results show that the variation of all five upregulated miRNAs was significantly higher in microdissected tumor cells compared with microdissected stromal cells (P<0.05), whereas only the results of two out of four downregulated miRNAs are statistically significant in microdissected tumor cells versus stromal cells (P<0.05). These results sharpen the significance of miRNA expression in tumor cells versus stromal cells (data not shown).
Putative Target Genes and their Potential Functions
We used several online software programs to investigate the prediction of the target genes, such as DIANA LAB, PicTar, miRDB and TargetScan. We took into consideration genes, which have been previously indentified, as differentially expressed between small intestinal neuroendocrine tumor cells and normal enterochromaffin cells.12 The match between miRNA sequences and mRNAs generated a list of potential target genes and the potential gene function has been considered. The results of this investigation are summarized in Supplementary Tables 2 and 3.
Discussion
MiRNAs are small RNAs, which regulate gene expression at post-transcriptional level. They are virtually involved in all the physiological processes, such as development, differentiation, heart function, metabolism, hemostasis and apoptosis. During the past 20 years numerous studies collected sufficient evidence to suggest that miRNAs are involved in a range of diseases, including cardiovascular disorders and cancer. Evidences have been reported about aberrant miRNA expression levels in tissues and in sera from patients with different forms of malignant tumors.28, 29
We investigated miRNA expression in well-differentiated small intestinal neuroendocrine tumors and our main goal was to provide a list of differentially expressed miRNAs as a source of novel potential biomarkers. Although most types of human cancers have been miRNA profiled,13, 17 miRNA expression in small intestinal neuroendocrine tumors has been not evaluated clearly.22 Thus, we used two different strategies of investigation to achieve this goal. The first one aimed at investigating potential differences in miRNA expression between primary tumors, and mesenteric and liver metastases, which may have an impact on tumor progression by using global miRNA array analysis, whereas the second goal aimed at identifying miRNAs, which showed relatively specific expression in immuno-laser capture microdissected normal enterochromaffin cells and laser capture microdissected small intestinal neuroendocrine tumor cells at different stages of disease.
Although we started with raw data coming from a global analysis, we filtered the number of significant miRNAs, which are differentially expressed in primary tumors versus mesentery and liver metastases, down to 33 for further analysis. Moreover, an advanced bioinformatics clustering approach has shown that 33 differentially expressed miRNAs clustered in a specific manner according to different stages of disease. Indeed, 5 primary tumors cluster together, whereas 9 out of 10 metastatic miRNAs cluster in an independent manner. MiRNAs clustering supports our previously published data,12 which has underlined the limited variation in gene expression between mesentery metastases versus liver metastases. These main findings show miRNAs’ capacity to classify the stage of small intestinal neuroendocrine tumors.
The selection of 33 miRNAs took into consideration the expression levels in tumor materials versus normal enterochromaffin cells, as well as the potential biological functions correlated to cell growth, apoptosis, differentiation and signal transduction. We paid major attention to the G-protein-coupled receptors (GPCRs) signaling transducer, underlining the importance of these receptors in endocrine and neuroendocrine cells30, 31, 32, 33 in terms of signal-transduction mechanisms. We have previously investigated GPCRs, such as the olfactory receptor 51 E1 (OR51E1), which has been reported as a potential novel carcinoma marker of small intestinal neuroendocrine tumors, as its restriction in terms of mRNA expression in normal enterochromaffin cell and tumor cells compared with a variety of different normal tissues.12 Moreover, we have presented unpublished novel data on OR51E1 as a potential novel tissue biomarker for the diagnosis and prognosis of small intestinal neuroendocrine tumors.34, 35 These novel findings address the potential importance of OR51E1 in small intestinal neuroendocrine tumors for further therapeutic strategies.
Furthermore, we focused on miRNAs, which were not previously associated to small intestinal neuroendocrine tumors to identify potential novel biomarkers. We then critically compared our results with previously published data by the RV Lloyd group,22 which are of outmost importance to compare our first global miRNA array expression analysis. We also realized that the selected group of miRNAs was distinct from the findings of different groups.36, 37, 38
A well-designed investigation triggered the most important findings of this study. First, the global miRNA profiling of small intestinal neuroendocrine tumors, then the validation of the filtered and selected miRNAs on tissue blocks, previously used for the profiling, followed by the miRNA expression of the selected miRNAs on immune laser capture microdissected normal enterochromaffin cells and laser capture microdissected small intestinal neuroendocrine tumor cells at different stages of disease, by using two independent test groups to extract microdissected cells. Moreover, we increased the reliability of the final results of this novel study introducing nine different miRNAs. The analysis of the selected miRNAs, following the Gene Ontology potential information, has been moving from tissue specimens to microdissected normal and tumor cells to exclude contaminant cells in the significance of differentially upregulated five miRNAs (miR-96, -182, -183, -196a and -200a) at different stages of disease and four (miR-31, -129-5p, -133a and -215) downregulated miRNAs. Indeed, it is of outmost importance to underline that these results were in full agreement by using two different test groups’ analyses. Moreover, we have shown the large variation in terms of miRNA expression between the microdissected primary tumor cells and the microdissected adjacent stromal cells.
Several analyses show that cancer networks are connected by miRNAs and the rationale for targeting miRNAs relies on two previous major findings. First, miRNA expression is deregulated in cancer cells compared with normal cells and the second one is that miRNA-based therapeutic approaches can be used to target specific protein expression.13 The ongoing research aims at screening established human neuroendocrine tumor cell lines, such as CNDT2.5 and KRJ-1, which are small intestinal neuroendocrine tumor cellular models, QGP-1 for pancreatic endocrine tumors and NCI-H720 and NCI-H727 for lung carcinoids to unveil miRNA targets effects on cell biology, that is, mRNA downregulation and target protein levels alteration. In conclusion, five miRNAs are significantly upregulated (miR-96, -182, -183, -196a and -200a), whereas four are significantly downregulated (miR-31, -129-5p, -133a and -215) during tumor progression. In addition, our results are in full agreement with data published previously.22 Nevertheless, we report here for the first time the variation of miR-31, -96, 129-5p, -182, -196a, -200a and -215 expressions in human small intestinal neuroendocrine tumors. In addition, the statistical significance of our results is undeniable and thus the selected miRNAs may be potentially developed either as prognostic or therapeutic targets in our opinion. Overall, miRNAs represent an important class of small RNAs with a huge impact on health and disease.
Future studies will further illuminate the potential value of miRNAs in diagnostics and therapeutics. Indeed, the future final goals are mRNA targets’ identification and miRNAs’ development as novel serum/plasma markers39 to create less invasive tests to monitor small intestinal neuroendocrine tumors. Although further experimental investigations are mandatory to achieve mRNA target recognition and blood test assay development, our study offers a new potential window to understand the role of specifically deregulated miRNAs in small intestinal neuroendocrine tumors at different stages of the disease at the best and potentially develop them as novel small intestinal neuroendocrine tumors’ clinical biomarkers. In conclusion, miRNAs may be used as biomarkers for small intestinal neuroendocrine tumors as they are valuable diagnostic tools.
Accession codes
References
Bosman TF, Carneiro F, Hruban RH et al(eds). WHO Classification of Tumours of the Digestive System. IARC Press: Lyon, 2010, p 13.
Rindi G, Falconi M, Klersy C et al TNM staging of neoplasms of the endocrine pancreas: results from a large international cohort study. J Natl Cancer Inst 2012;104:764–777.
Essand M, Leja J, Giandomenico V et al Oncolytic viruses for the treatment of neuroendocrine tumors. Horm Metab Res 2011;43:877–883.
Rindi G, Wiedenmann B . Neuroendocrine neoplasms of the gut and pancreas: new insights. Nat Rev Endocrinol 2012;8:54–64.
Cunningham JL, Diaz de Stahl T, Sjoblom T et al Common pathogenetic mechanism involving human chromosome 18 in familial and sporadic ileal carcinoid tumors. Genes Chromosomes Cancer 2011;50:82–94.
Cunningham JL, Janson ET . The biological hallmarks of ileal carcinoids. Eur J Clin Invest 2011;41:1353–1360.
Essand M, Vikman S, Grawe J et al Identification and characterization of a novel splicing variant of vesicular monoamine transporter 1. J Mol Endocrinol 2005;35:489–501.
Leja J, Dzojic H, Gustafson E et al A novel chromogranin-A promoter-driven oncolytic adenovirus for midgut carcinoid therapy. Clin Cancer Res 2007;13:2455–2462.
Leja J, Yu D, Nilsson B et al Oncolytic adenovirus modified with somatostatin motifs for selective infection of neuroendocrine tumor cells. Gene Therapy 2011;18:1052–1062.
Vikman S, Giandomenico V, Sommaggio R et al CD8+ T cells against multiple tumor-associated antigens in peripheral blood of midgut carcinoid patients. Cancer Immunol Immunother 2008;57:399–409.
Cui T, Hurtig M, Elgue G et al Paraneoplastic antigen Ma2 autoantibodies as specific blood biomarkers for detection of early recurrence of small intestine neuroendocrine tumors. PLoS One 2010;5:e16010.
Leja J, Essaghir A, Essand M et al Novel markers for enterochromaffin cells and gastrointestinal neuroendocrine carcinomas. Mod Pathol 2009;22:261–272.
Garzon R, Marcucci G, Croce CM . Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 2010;9:775–789.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297.
Bartel DP, Chen CZ . Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5:396–400.
Sakamoto S, Aoki K, Higuchi T et al The NF90–NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol 2009;29:3754–3769.
Zhong X, Coukos G, Zhang L . miRNAs in human cancer. Methods Mol Biol 2012;822:295–306.
Brown BD, Naldini L . Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet 2009;10:578–585.
Lee HW, Lee EH, Ha SY et al Altered expression of microRNA miR-21, miR-155, and let-7a and their roles in pulmonary neuroendocrine tumors. Pathol Int 2012;62:583–591.
Markou A, Liang Y, Lianidou E . Prognostic, therapeutic and diagnostic potential of microRNAs in non-small cell lung cancer. Clin Chem Lab Med 2011;49:1591–1603.
Voortman J, Lee JH, Killian JK et al Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci USA 2010;107:13040–13045.
Ruebel K, Leontovich AA, Stilling GA et al MicroRNA expression in ileal carcinoid tumors: downregulation of microRNA-133a with tumor progression. Mod Pathol 2010;23:367–375.
Gao Y, Schug J, McKenna LB et al Tissue-specific regulation of mouse microRNA genes in endoderm-derived tissues. Nucleic Acids Res 2011;39:454–463.
Saeed AI, Bhagabati NK, Braisted JC et al TM4 microarray software suite. Methods Enzymol 2006;411:134–193.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 2001;25:402–408.
Nakamura N, Ruebel K, Jin L et al Laser capture microdissection for analysis of single cells. Methods Mol Med 2007;132:11–18.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20.
Allegra A, Alonci A, Campo S et al Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (Review). Int J Oncol 2012;;41:1897–1912.
You JS, Jones PA . Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 2012;22:9–20.
Braun T, Voland P, Kunz L et al Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology 2007;132:1890–1901.
Leu FP, Nandi M . GPCR somatostatin receptor extracellular loop 2 is a key ectodomain for making subtype-selective antibodies with agonist-like activities in the pancreatic neuroendocrine tumor BON cell line. Pancreas 2010;39:1155–1166.
Millar RP, Newton CL . The year in G protein-coupled receptor research. Mol Endocrinol 2010;24:261–274.
Woehler A, Ponimaskin EG . G protein-mediated signaling: same receptor, multiple effectors. Curr Mol Pharmacol 2009;2:237–248.
Kaku M, Nishiyama T, Yagawa K et al Establishment of a carcinoembryonic antigen-producing cell line from human pancreatic carcinoma. Gann 1980;71:596–601.
Cui T, Tsolakis AV, Cunningham J et al Olfactory receptor 51E1 is a potential novel tissue biomarker for the diagnosis and prognosis of small intestine neuroendocrine tumors. Regul Pept 2012;177:S18.
Hamfjord J, Stangeland AM, Hughes T et al Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One 2012;7:e34150.
Heneghan HM, Miller N, Kerin MJ . MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol 2010;10:543–550.
Roldo C, Missiaglia E, Hagan JP et al MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol 2006;24:4677–4684.
Cortez MA, Bueso-Ramos C, Ferdin J et al MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011;8:467–477.
Acknowledgements
We thank Professor Magnus Essand for critical reading of the manuscript. We also thank Åsa Forsberg and Dr Jan Grawé for excellent technical assistance and David Brodin for critical assistance with the microRNA array analysis. This work was supported by funding from the Lions Cancerforskningsfond, Tore Nilssons Stiftelse för Medicinsk Forskning and Erik, Karin och Gösta Selander Stiftelse. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Li, SC., Essaghir, A., Martijn, C. et al. Global microRNA profiling of well-differentiated small intestinal neuroendocrine tumors. Mod Pathol 26, 685–696 (2013). https://doi.org/10.1038/modpathol.2012.216
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.216
Keywords
This article is cited by
-
Molecular Pathology of Well-Differentiated Gastro-entero-pancreatic Neuroendocrine Tumors
Endocrine Pathology (2021)
-
Exosomal miRNA signatures of pancreatic lesions
BMC Gastroenterology (2020)
-
Treatment with somatostatin analogs induces differentially expressed let-7c-5p and mir-3137 in small intestine neuroendocrine tumors
BMC Cancer (2019)
-
Role of biomarker tests for diagnosis of neuroendocrine tumours
Nature Reviews Endocrinology (2018)
-
miRNA profiling of small intestinal neuroendocrine tumors defines novel molecular subtypes and identifies miR-375 as a biomarker of patient survival
Modern Pathology (2018)