Abstract
The HER2 oncogene was recently reported to be amplified and overexpressed in esophageal adenocarcinoma. However, the relationship of HER2 amplification in esophageal adenocarcinoma with prognosis has not been well defined. The scoring systems for clinically evaluating HER2 in esophageal adenocarcinoma are not established. The aims of the study were to establish a HER2 scoring system and comprehensively investigate HER2 amplification and overexpression in esophageal adenocarcinoma and its precursor lesion. Using a tissue microarray, containing 116 cases of esophageal adenocarcinoma, 34 cases of Barrett's esophagus, 18 cases of low-grade dysplasia and 15 cases of high-grade dysplasia, HER2 amplification and overexpression were analyzed by HercepTest and chromogenic in situ hybridization methods. The amplification frequency in an independent series of 116 esophageal adenocarcinoma samples was also analyzed using Affymetrix SNP 6.0 microarrays. In our studies, we have found that HER2 amplification does not associate with poor prognosis in total 232 esophageal adenocarcinoma patients by chromogenic in situ hybridization and high-density microarrays. We further confirm the similar frequency of HER2 amplification by chromogenic in situ hybridization (18%; 21 out of 116) and SNP 6.0 microarrays (16%, 19 out of 116) in esophageal adenocarcinoma. HER2 protein overexpression was observed in 12% (14 out of 116) of esophageal adenocarcinoma and 7% (1 out of 15) of high-grade dysplasia. No HER2 amplification or overexpression was identified in Barrett's esophagus or low-grade dysplasia. All HER2 protein overexpression cases showed HER2 gene amplification. Gene amplification was found to be more frequent by chromogenic in situ hybridization than protein overexpression in esophageal adenocarcinoma (18 vs 12%). A modified two-step model for esophageal adenocarcinoma HER2 testing is recommended for clinical esophageal adenocarcinoma HER2 trial.
Similar content being viewed by others
Main
Esophageal adenocarcinoma is thought to develop as a result of gastroesophageal reflux, which initiates a metaplastic change in the lower esophageal epithelium. Accumulation of genetic and epigenetic changes then results in progression to dysplasia and cancer in some individuals.1, 2 The incidence of esophageal adenocarcinoma has increased rapidly in the last three decades and the prognosis is usually very poor with 5-year survival rates ranging from 14–22%.3, 4, 5, 6 The surgical treatment of esophageal adenocarcinoma can offer cure for some, however, many patients have locally advanced or disseminated disease at presentation and require systemic therapy. Current chemotherapy regimens provide only minimal survival benefit, predominantly when used in combination with surgery or radiation. Identification of genetic alterations in esophageal adenocarcinoma that offer potential for biologically targeted treatment is one of the best hopes to improve the selectivity of therapy and enhance patient survival.
The HER2 (c-erbB2) gene, a proto-oncogene, is located on chromosome 17q11.2-12 and encodes epidermal growth factor receptor 2 (HER2), a transmembrane glycoprotein receptor p185HER2, which is targeted by the humanized monoclonal antibody trastuzumab (Herceptin).7 HER2 is amplified and overexpressed in ∼25% of breast cancer patients and is associated with an aggressive clinical course and poor prognosis.8 Trastuzumab treatment in combination with chemotherapy for breast cancer patients with HER2 amplification/overexpression has shown a significant clinical efficacy in both the metastatic and adjuvant settings.9 In esophageal adenocarcinoma, HER2 overexpression and amplification has been reported at frequencies similar to those observed in breast cancer.8, 10, 11, 12, 13, 14, 15, 16 In esophageal adenocarcinoma, however, the relationship between HER2 amplification and prognosis is controversial.12, 17 Recent studies have shown that anti-HER2 treatment enhanced radiosensitivity of esophageal cancer cell lines with HER2 overexpression.18 In esophageal adenocarcinoma, although the initial results from 19 esophageal adenocarcinoma patients did not show significant improvement of patient survival,19 ToGA clinical trials in patients with gastric adenocarcinoma (trial vs control: 236 vs 243 patients) and gastroesophageal junction adenocarcinoma (trial vs control: 58 vs 48 patients) have shown a significant survival benefit for patients treated with a combination of Trastuzumab and standard chemotherapy.20, 21 The interest for treatment of esophageal adenocarcinoma therefore remains high.
The guidelines for detection and interpretation of HER2 amplification/overexpression in breast cancer have been well established and anti-HER2 targeted treatment is routinely considered in the management of these patients.22 Recently, Hofmann et al23 established a modified HER2 scoring system in gastric cancer to identify suitable patients for enrollment in clinical trastuzumab trial. They found an incomplete moderate to strong basal/lateral membranous HER2 staining pattern and higher rate of tumor heterogeneity in gastric adenocarcinoma compared with breast cancer. However, the clinical assessment of HER2 amplification and overexpression with immunohistochemistry and chromogenic in situ hybridization in esophageal adenocarcinoma is not well defined. In addition, the data on the frequency of HER2 amplification and overexpression in esophageal adenocarcinoma and its precursor lesions including columnar cell metaplasia, Barrett's esophagus and dysplasia is very limited, particularly in US populations. Therefore, the aims of this study are (1) to comprehensively investigate the frequency of HER2 overexpression and amplification in esophageal adenocarcinoma and precursor lesion using HER2 immunohistochemistry, chromogenic in situ hybridization and SNP 6.0 microarray, (2) to establish clinical methods to assess HER2 amplification and overexpression, (3) to determine whether HER2 amplification is a prognostic marker for esophageal adenocarcinoma patients by chromogenic in situ hybridization and high definition microarray in separate large clinical specimen and (4) to determine the relationship of HER2 amplification with clinical factors including sex, age, lymph node metastasis and tumor stage.
Materials and methods
Construction of Tissue Microarray
Tissue microarrays, containing 38 cases of Barrett's esophagus, 81 cases of columnar cell metaplasia, 86 cases of squamous epithelium, 18 cases of low-grade dysplasia, 15 cases of high-grade dysplasia, and 116 cases of esophageal adenocarcinoma, were constructed from the representative areas of formalin-fixed specimens collected between 1997–2005 in the Department of Pathology and Laboratory Medicine, University of Rochester Medical Center/Strong Memorial Hospital, Rochester, New York. The 5-μ sections were cut from tissue microarrays and were stained with H&E to confirm the presence of the expected tissue histology within each tissue core. Additional sections were cut for immunohistochemistry and chromogenic in situ hybridization analysis.
Patients for Tissue Microarrays
All the 116 patients with esophageal adenocarcinoma used for the tissue microarray construction were treated with esophagectomy in Strong Memorial Hospital/University of Rochester between 1997 and 2005. These patients included 104 males (90%) and 12 females (10%). The patient age ranged from 34 to 85 years with a mean of 65 years. The follow-up period after esophagectomy ranged from 0.03 to 142 months with a mean of 39 months.
Patients for Affymetrix SNP 6.0 Analysis
Frozen tumors were obtained from 116 patients undergoing esophagectomy at the University of Pittsburgh Medical Center, Pittsburgh, PA between 2002 and 2008. Patient age ranged from 43–88 and the cohort consisted of 95 males and 21 females. Final pathologic stages were stage I (28), stage II (31), stage III (49) and stage IV (7). All tumor specimens were evaluated by a pathologist and were determined to be >70% tumor cell representation. Further details of this patient cohort and comprehensive genomic analysis of these tumors is to be published elsewhere.
In both institutes, all research was performed under protocols approved at both participating institutions.
Affymetrix SNP 6.0 Analysis
Genomic DNA was isolated using the QiaAmp DNA Mini Kit (Qiagen, CA) and 600 ng was used for labeling and array hybridization at the SUNY Upstate Medical University microarray core facility (Syracuse, NY) using kits and protocols provided by Affymetrix. Array data quality was assessed using Affymetrix Genotyping Console 3.0 and all further data analysis was performed using Nexus 5.0 Copy Number Analysis software (Biodiscovery, CA).
Immunohistochemistry
Tissue sections from the tissue microarray were deparaffinized, rehydrated through graded alcohols, and washed with phosphate buffered saline. Antigen retrieval for HER2 was performed by heating sections in 99 °C water bath for 40 min. After endogenous peroxidase activity was quenched and nonspecific binding was blocked, ready-to-use mouse monoclonal antibody anti-HER2 (DAKO, CA) was incubated at room temperature for 30 min. The secondary antibody (Flex HRP) was allowed to incubate for 30 min. After washing, sections were incubated with Flex DAB chromogen for 10 min and counterstained with Flex hematoxylin for 5 min. A breast carcinoma with known HER2 overexpression served as positive control. Negative control was performed by replacing anti-HER2 antibody with normal serum.
Chromogenic In Situ Hybridization
HER2 chromogenic in situ hybridization was performed according to the manufacturer's protocol (DAKO DuoCISH kit, DAKO, CA) and stained in the Dako Autostainer. Briefly, tissue microarray sections were deparaffined in xylene, rehydrated through graded alcohols and washed in diluted wash buffer for 3 min. Endogenous peroxidase was blocked by peroxidase block solution for 5 min. Sections were subsequently incubated with chromogenic in situ hybridization antibody mix for 30 min. After washing, sections were incubated with red chromogen solution and blue chromogen solution for 10 min, respectively. Sections were counterstained with hematoxylin, dried and mounted. A breast carcinoma and esophageal adenocarcinoma with known HER2 amplification was used as positive control. Negative control was performed by replacing HER2 probe by normal serum.
Scoring of Immunohistochemistry and Chromogenic In Situ Hybridization
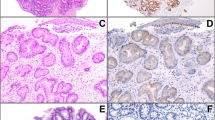
All sections were reviewed independently by ZZ and YH blinded to all clinical and pathologic information. Discordant cases were reviewed by DH and a final consensus was reached. For HER2 immunohistochemical stain, percentage (0-100%) of positive cells was determined. Gastric HER2 scoring criteria were used for evaluation of these samples. The intensity of HER2 staining was graded as 0, 1+, 2+, or 3+. HER2 protein was considered overexpressed if 10% or more of cells stained with a moderate to strong intensity and showed either complete or a basal/lateral membrane staining pattern (Figure 3, Table 4). The luminal part of well to moderately differentiated tumor glands often show negative stain. This pattern is also observed in gastric adenocarcinoma.23 For HER2 chromogenic in situ hybridization, the hybridization signals were counted in 50 nuclei per tissue core (Figure 2). All overlapping nuclei were excluded, only nuclei with a distinct nuclear border were evaluated. The ratio between HER2 and chromosome 17 centromere copy numbers was calculated. The HER2 gene was considered as amplification when the ratio of gene-specific HER2: centromere probe signals (CEP17) was ≥2.0.
Statistical analysis
Kaplan–Meier survival estimator and logrank test was used to analyze the patient survival between HER2 amplified group and non-HER2 amplified group. Both a univariate model with HER2 as the soul explanatory variable and a multivariate model with several other clinical covariate, including gender, age, lymph node metastasis and tumor stage, were used. In addition, Fisher's exact test and χ2-test were used to compare HER2 positivity rate between esophageal adenocarcinoma, high- and low-grade dysplasia, columnar cell metaplasia and squamous epithelium subpopulations. P-value less than 0.05 is considered statistically significant.
Results
Defining the HER2/ERBB2 Amplicon in Esophageal Adenocarcinoma With High-Density Microarrays
Analysis of 116 esophageal adenocarcinoma specimens using high-density microarrays revealed amplification in 16% (19 out of 116). In 9% (10 out of 116), amplification was considered high level (log2 ratio >0.6). The minimal region of amplification spans 68 kb (Figure 1) and contains only three genes (ERBB2, C17orf37 and GRB7). In this cohort study, the median overall survival of HER2 amplification is 21 months and non-HER2 amplification is 25 months. No association was found with HER2 amplification and either disease free survival (P=0.709) or overall survival (P=0.27).
Frequency histogram showing amplification of the ERBB2 (HER2) locus at chromosome 17q12 in 116 esophageal adenocarcinoma samples. The minimal amplicon (peak) spans 68 kb and contains only three genes, ERBB2, C17orf37 and GRB7. This locus is amplified in 19 out of 116 (16%) cases in this patient cohort.
Chromogenic In Situ Hybridization Analysis
The HER2 chromogenic in situ hybridization results are summarized in Table 1. No HER2 gene amplification was identified in esophageal squamous epithelium (n=86), columnar cell metaplasia (n=81), Barrett's esophagus (n=34) and low-grade dysplasia (n=18). One of 15 cases of high-grade dysplasia (7%) showed HER2 gene amplification (Table 1) and 21 of 116 cases of esophageal adenocarcinoma (18.10%) showed HER2 gene amplification (Table 1, Figure 2). Esophageal adenocarcinoma showed statistically significant higher HER2 gene amplification compared with the low-grade dysplasia, Barrett's esophagus, columnar cell metaplasia, and squamous epithelium groups (P<0.05), but not when compared with high-grade dysplasia group (P>0.05).
Immunohistochemical Analysis
The HER2 immunohistochemical staining results are summarized in Table 1 and Figure 3. No HER2 protein overexpression was identified in esophageal squamous epithelium (n=86), columnar cell metaplasia (n=81), Barrett's esophagus (n=34) and low-grade dysplasia (n=18). One of 15 cases of high-grade dysplasia (7%) showed HER2 overexpression (Table 1). Eleven of 116 cases of esophageal adenocarcinoma (10%) showed HER2 overexpression from tissue microarray (Table 1, Figure 3). The whole slides from 10 cases with HER2 amplification and non-HER2 overexpression were used to investigate the heterogeneity of HER2 overexpression. Three cases in whole slides showed focal positive immunohistochemical stain (2+). Total 14 of 116 cases of esophageal adenocarcinoma (12%) showed HER2 overexpression. esophageal adenocarcinoma showed statistically significant higher HER2 protein overexpression compared with the low grade dysplasia, Barrett's esophagus, columnar cell metaplasia, and squamous epithelium groups (P<0.05), but not when compared with high-grade dysplasia group (P>0.05).
HER2 immunohistochemical staining showing 0 immunostaining ((a) × 400), 1+ immunostaining ((b) × 400), 2+ immunostaining ((c) × 400) and 3+ immunostaining ((d) × 400) in esophageal adenocarcinoma. Both 2+ and 3+ uniform staining are considered as HER2 protein overexpression. Incomplete membranous ‘U’ shape stain is presented in c.
Association Between Immunohistochemistry and Chromogenic In Situ Hybridization Analyses
The correlation between HER2 protein overexpression by immunohistochemical stain and HER2 gene amplification by chromogenic in situ hybridization in 116 cases of esophageal adenocarcinoma is summarized in Table 2. All 14 cases of esophageal adenocarcinoma with HER2 protein overexpression identified by immunohistochemical stain also showed HER2 gene amplification by chromogenic in situ hybridization. Ten cases with HER2 gene amplification identified by chromogenic in situ hybridization showed HER2 protein overexpression in three cases with whole slides immunohistochemical stain (Table 2). No HER2 expression showed in the other seven cases. Total 95 cases (82%) of esophageal adenocarcinoma showed neither HER2 protein overexpression nor HER2 gene amplification. Concordance between immunohistochemical stain and chromogenic in situ hybridization was 76%.
Association of HER2 Amplification with Survival and Other Clinical Factors
The median survival after esophagectomy between HER2 amplification group and no HER2 amplification group is 25 and 23 months, respectively, by chromogenic in situ hybridization. A survival analysis was based on Cox regression and log-rank test showed that there is no statistical significance in overall survival between the two groups (P=0.19, Figure 4). When this regression was refit by using one of the following clinical covariate: age, gender, the stage, and metastatic lymph nodes of the tumor, instead of HER2, we found that age (P=0.064) and gender (P=0.448) were not significantly associated with patient survival, but the stage (P<0.001) and the number of metastatic lymph nodes of the tumor (P<0.001) have strong association with patient survival.
In addition, we studied the association between HER2 amplification and these clinical factors (Table 3). Of 116 esophageal adenocarcinoma patients, 21 had HER2 amplification. Nineteen were male, and two female (M:F ratio, 10:1), with a mean age of 63 years (range, 51 to 74 years). The remaining patients (85 males and 10 females (M:F ratio, 9:1), mean age 85 years, (34–85 years)) had no amplification. A Fisher's exact test shows that there is no significant association between HER2 and gender (P=1.0), age (P=0.188), the stage (P=0.325), and the number of metastatic lymph nodes (P=0.234). However, the frequency of HER2 amplification was found to be significantly higher (P=0.004) in moderately differentiated tumors (13/22) compared with poor or well-differentiated tumors (1/6 and 7/61 respectively).
Discussion
In the present study, we provide evidence that HER2 amplification does not associate with poor prognosis in total 232 esophageal adenocarcinoma patients by chromogenic in situ hybridization (116 patients) and high-density microarrays (116 patients). We further confirm the frequency of HER2 amplification and overexpression in esophageal adenocarcinoma and high-grade dysplasia (see Table 1) by various methods including high-density microarrays, chromogenic in situ hybridization and immunohistochemistry. In addition, we recommend a modified two-step model for clinical HER2 study in esophageal adenocarcinoma patients, similar to what has been proposed for the HER2 evaluation of gastric adenocarcinomas.23
Data on the association between HER2 amplification and survival duration in esophageal adenocarcinoma are limited and conflicting. Brien et al17 found that patients with HER2 amplification (n=11) had shorter survival durations than did patients without amplification (n=43). However, Reichelt et al12 found no survival difference between the HER2 amplification (n=16) and no HER2 amplification groups (n=90)(P=0.953). In addition, Rauser et al24 found that HER2 gene amplification was associated with increased disease-specific mortality on three-dimensional fluorescence in situ hybridization (FISH) analysis in thick slides (16 μm), but not on FISH and immunohistochemical analyses in thin (4 μm) sections. Our results indicate no association of HER2 amplification with patient survival in a large cohort studies (total 232 patients) by both chromogenic in situ hybridization and high-density microarrays.
In esophageal adenocarcinoma, recently Reichelt et al12 found that 15% (16 out of 110) of tumors had HER2 gene amplification with FISH. Similarly, Brien et al. showed that 19% (12 out of 63) of esophageal adenocarcinomas had HER2 gene amplification.17 In addition, with three-dimentional FISH method in thick slides (16 μm, n=124), Rauser et al24 found that HER2 amplification was 10.5% in high-level amplification (≥6.0 signals) and 60% in low-level copy number change (≥2.5–4.0 signals). However, in thin slides (4 μm, n=123), HER2 amplification was found in 9% in high-level amplification (≥6.0 signals) and 6% in low-level copy number change (≥2.5–4.0 signals). In the current study, we found that HER2 amplification was 18% (21 out of 116) detected by chromogenic in situ hybridization and 16.4% (19 out of 116) by high definition microarray in cases of esophageal adenocarcinoma. In addition we found no evidence of HER2 amplification in low-grad dysplasia, Barrett's esophagus, columnar cell metaplasia or normal esophageal squamous epithelium. Thus, the frequency of HER2 amplification in esophageal adenocarcinoma appears to be consistent between studies with a range of 15–19% and this event appears not to occur before the development of high-grade dysplasia. However, there is a huge difference between traditional FISH in thin section (6%) and three-dimensional FISH in thick section (60%) to detect the low-level HER2 amplification. They considered that the tumor cell nuclei were truncated due to standardized thin tissue sectioning. Therefore, three dimension FISH need to be further evaluated to help better understand any prognostic significance.
With previous gene amplification with PCR and microarray analysis, the 17q gain was found in various percentage from 21–33% in Albrecht et al25 (n=18), van Dekken et al26 (n=28) and Miller et al27 (n=87). In current studies with high definition microarray analysis, 16.4% (19 out of 116) had HER2 amplification detected. The incidences of amplification defined by high-density microarrays in our study is close to reported FISH and chromogenic in situ hybridization results, but lower than those of previous studies.
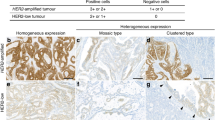
The guidelines for detection and interpretation of HER2 amplification/overexpression in breast cancer have been well established and anti-HER2 targeted treatment is routinely considered in the management of these patients.22, 28, 29, 30 Recently, HER2 scoring for gastric cancer has been modified due to HER2 stain pattern and heterogeneity,17 which helps to improve the concordance between immunohistochemical stain and FISH and will help select the most appropriate HER2 positive patients for clinical trials with a combination of Trastuzumab and standard chemotherapy. The guideline for HER2 scoring in esophageal adenocarcinoma is critical for enrolling the patient for clinical trial. In our study, all HER2 with 2+ and 3+ expression cases identified with immunohistochemical stain showed HER2 amplification with chromogenic in situ hybridization. The result is similar to the results of Reichelt's study from Germany,12 but it does not agree to the gastric carcinoma studies.23 With 2+ expression of HER2, only 36% of immunohistochemical stain 2+ cases were also FISH positive in gastric carcinoma. There are many causes of disagreement including immunohistochemical stain reactivity of esophageal adenocarcinoma, antibody, immunohistochemical methods, tissue fixation condition and etc. Actually, the HercepTest kit (DACO) was used in both studies. In addition, our studies were repeated several times in different immunohistochemical methods for the best results. In our study, modified two-step model is recommended for the guideline of clinical HER2 study in esophageal adenocarcinoma. In this model, immunohistochemical stain is first used to detect HER2 overexpression with 2+ and 3+ intensity and >10% of positive cells (see Table 4). If HER2 protein is overexpressed, HER2 DNA amplification can be assumed, and chromogenic in situ hybridization analysis may not be needed. If HER2 protein is not overexpressed, chromogenic in situ hybridization or FISH analysis should be performed to rule out gene amplification. The HER2 gene was considered as amplification when the ratio of gene-specific:centromere probe signals was ≥2.0 in our study, which is also the criteria used by Hofmann et al.23 The definition for HER2 overexpression should be 2+ and 3+ instead of 3+ only in breast and gastric cancer as all 2+ expression cases in esophageal adenocarcinoma showed HER2 amplification by chromogenic in situ hybridization. The negative overexpression of HER2 in esophageal adenocarcinoma also need to do chromogenic in situ hybridization as 7 of 102 patients show positive for chromogenic in situ hybridization test with negative for immunohistochemical HER2 overexpression (1+ or 0). In addition, the ‘U’ shaped stain pattern in esophageal adenocarcinoma without complete membrane is the common pattern in well differentiated esophageal adenocarcinoma, which is considered as positive immunohistochemical stain. The heterogeneity of HER2 overexpression in gastric adenocarcinoma is higher than that in breast cancer.23 We also found that the heterogeneity in esophageal adenocarcinoma is in our preliminary data in 10 chromogenic in situ hybridization positive and immunohistochemical negative cases. There is a potential problem to evaluate HER2 overexpression of the biopsy specimen in clinical practice. The heterogeneity will be further studied in following research. For gastric biopsy, Hofmann et al23 suggested that immunohistochemical intensity 3+ or FISH positive gastric adenocarcinoma should be counted as positive even in less than 10% cells. However, comparison of the treatment response from patients with less than 10% HER2 overexpression or amplification vs more than 10% should be studied. At present, we recommended that multiple biopsies of esophageal adenocarcinoma and gastric adenocarcinoma should be tested for HER2 amplification and overexpression. The criteria for gastric biopsy can be used for esophageal biopsy.
For our study, we used chromogenic in situ hybridization instead of FISH because of the increased stability of the chromogenic in situ hybridization labeled slides and the ease of interpretation of results, which does not require a fluorescent microscope and dark room. Furthermore the use of a light microscope allows us to see both signals and morphology simultaneously. Our experience and that of other groups showed that chromogenic in situ hybridization results are equivalent to FISH and that chromogenic in situ hybridization is a reliable method for detecting HER2 genomic abnormalities.31, 32 Although both have similar turnaround time and cost in the laboratory, the DNA-specific probes for chromogenic in situ hybridization are 25–50% cheaper than those for FISH.31 Because of the disadvantages of FISH, chromogenic in situ hybridization has shown a great potential to replace FISH for detecting HER2 gene amplificaton.32 In addition, our results showed that chromogenic in situ hybridization is more sensitive than immunohistochemical stain to detect HER2 abnormalities. This may be because DNA is more stable than protein during the specimen processing. However, the amplification without overexpression in some tumors also was reported.12
In summary, our study showed that HER2 is overexpressed at the protein level and amplified at the DNA level in a subset of esophageal adenocarcinoma and high-grade dysplasia. We observed neither HER2 protein overexpression nor DNA amplification in normal esophageal squamous epithelium, columnar cell metaplasia, Barrett's esophagus or low-grade dysplasia. A modified two-step model for the evaluation of the HER2 status in esophageal adenocarcinoma is suggested by our studies. Our results showed that HER2 amplification is not a prognostic predictor for esophageal adenocarcinoma. However, ToGA clinical trial for gastric and GE junctional adenocarcinoma with significant improving patient's survival suggests that the patients with HER2 positive esophageal adenocarcinoma would be potential candidates for anti-HER2 targeted treatment.
References
Bailey T, Biddlestone L, Shepherd N, et al. Altered cadherin and catenin complexes in the Barrett's esophagus-dysplasia-adenocarcinoma sequence: correlation with disease progression and dedifferentiation. Am J pathol 1998;152:135–144.
Maru DM, Singh RR, Hannah C, et al. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol 2009;174:1940–1948.
Gamliel Z, Krasna MJ . Multimodality treatment of esophageal cancer. Surg Clin North Am 2005;85:621–630.
Luketich JD, Alvelo-Rivera M, Buenaventura PO, et al. Minimally invasive esophagectomy: outcomes in 222 patients. Annals of surgery 2003;238:486–494;discussion 94–5.
Swanson SJ, Batirel HF, Bueno R, et al. Transthoracic esophagectomy with radical mediastinal and abdominal lymph node dissection and cervical esophagogastrostomy for esophageal carcinoma. Ann Thorac Surg 2001;72:1918–1924 discussion 24–5.
Pohl H, Welch HG . The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst 2005;97:142–146.
Baselga J, Tripathy D, Mendelsohn J, et al. Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol 1996;14:737–744.
Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987;235:177–182.
Goldenberg MM . Trastuzumab, a recombinant DNA-derived humanized monoclonal antibody, a novel agent for the treatment of metastatic breast cancer. Clin Ther 1999;21:309–318.
Rossi E, Villanacci V, Bassotti G, et al. Her-2/neu in barrett esophagus: a comparative study between histology, immunohistochemistry, and fluorescence in situ hybridization. Diagn Mol Pathol 2006;15:125–130.
Rossi E, Grisanti S, Villanacci V, et al. Her-2 overexpression/amplification in Barrett's esophagus predicts early transition from dysplasia to adenocarcinoma: A Clinico-Pathologic Study. J Cell Mol Med 2009;13:3826–3833.
Reichelt U, Duesedau P, Tsourlakis M, et al. Frequent homogeneous HER-2 amplification in primary and metastatic adenocarcinoma of the esophagus. Mod Pathol 2007;20:120–129.
Langer R, Von Rahden BH, Nahrig J, et al. Prognostic significance of expression patterns of c-erbB-2, p53, p16INK4A, p27KIP1, cyclin D1 and epidermal growth factor receptor in oesophageal adenocarcinoma: a tissue microarray study. J Clin Pathol 2006;59:631–634.
Dreilich M, Wanders A, Brattstrom D, et al. HER-2 overexpression (3+) in patients with squamous cell esophageal carcinoma correlates with poorer survival. Dis Esophagus 2006;19:224–231.
Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989;244:707–712.
Zhang D, Salto-Tellez M, Do E, et al. Evaluation of HER-2/neu oncogene status in breast tumors on tissue microarrays. Hum Pathol 2003;34:362–368.
Brien TP, Odze RD, Sheehan CE, et al. HER-2/neu gene amplification by FISH predicts poor survival in Barrett's esophagus-associated adenocarcinoma. Hum Pathol 2000;31:35–39.
Sato S, Kajiyama Y, Sugano M, et al. Monoclonal antibody to HER-2/neu receptor enhances radiosensitivity of esophageal cancer cell lines expressing HER-2/neu oncoprotein. Int J Radiat Oncol Biol Phys 2005;61:203–211.
Safran H, Dipetrillo T, Akerman P, et al. Phase I/II study of trastuzumab, paclitaxel, cisplatin and radiation for locally advanced, HER2 overexpressing, esophageal adenocarcinoma. Int J Radiat Oncol Biol Phys 2007;67:405–409.
Jorgensen JT . Target HER2 treatment in advanced gastric cancer. Oncology 2010;78:26–33.
Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687–697.
Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med 2007;131:18–43.
Hofmann M, Stoss O, Shi D, et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 2008;52:797–805.
Rauser S, Weis R, Braselmann H, et al. Significance of HER2 low-level copy gain in Barrett's cancer: implications for fluorescence in situ hybridization testing in tissues. Clin Cancer Res 2007;13:5115–5123.
Albrecht B, Hausmann M, Zitzelsberger H, et al. Array-based comparative genomic hybridization for the detection of DNA sequence copy number changes in Barrett's adenocarcinoma. J Pathol 2004;203:780–788.
van Dekken H, Geelen E, Dinjens WN, et al. Comparative genomic hybridization of cancer of the gastroesophageal junction: deletion of 14Q31-32.1 discriminates between esophageal (Barrett's) and gastric cardia adenocarcinomas. Cancer Res 1999;59:748–752.
Miller CT, Moy JR, Lin L, et al. Gene amplification in esophageal adenocarcinomas and Barrett's with high-grade dysplasia. Clin Cancer Res 2003;9:4819–4825.
Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 1999;17:2639–2648.
Joensuu H, Kellokumpu-Lehtinen PL, Bono P, et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med 2006;354:809–820.
Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007;25:118–145.
Mayr D, Heim S, Weyrauch K, et al. Chromogenic in situ hybridization for Her-2/neu-oncogene in breast cancer: comparison of a new dual-colour chromogenic in situ hybridization with immunohistochemistry and fluorescence in situ hybridization. Histopathology 2009;55:716–723.
Gong Y, Sweet W, Duh YJ, et al. Chromogenic in situ hybridization is a reliable method for detecting HER2 gene status in breast cancer: a multicenter study using conventional scoring criteria and the new ASCO/CAP recommendations. Am J Clin Pathol 2009;131:490–497.
Acknowledgements
We thank Dr Jorge Yao for the tissue microarray construction and Qi Yang for immunohistochemical stain and chromogenic in situ hybridization study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hu, Y., Bandla, S., Godfrey, T. et al. HER2 amplification, overexpression and score criteria in esophageal adenocarcinoma. Mod Pathol 24, 899–907 (2011). https://doi.org/10.1038/modpathol.2011.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2011.47
Keywords
This article is cited by
-
Optimal First-Line Therapy for Metastatic Adenocarcinoma of the Esophagus
Current Treatment Options in Oncology (2022)
-
Expression and significance of Her2 and Ki-67 in gastric adenocarcinoma without distant metastasis: a cohort study
BMC Gastroenterology (2020)
-
High expression of carbonic anhydrase IX is significantly associated with glandular lesions in gastroesophageal junction and with tumorigenesis markers BMI1, MCM4 and MCM7
BMC Gastroenterology (2015)
-
Systematic review and meta-analysis of immunohistochemical prognostic biomarkers in resected oesophageal adenocarcinoma
British Journal of Cancer (2015)
-
Determination of Tumor Heterogeneity in Colorectal Cancers Using Heterogeneity Tissue Microarrays
Pathology & Oncology Research (2015)