Abstract
The significance of KRAS mutant allele-specific imbalance (MASI) in lung adenocarcinomas is unknown. KRAS MASI was defined as predominance of the mutant allele over the wild-type allele. We assessed the frequency of KRAS MASI by comparing peak heights of mutant and wild-type alleles on sequencing electropherograms and by KRAS fluorescence in situ hybridization (FISH). A review of sequencing electropherograms of 207 KRAS-mutated lung adenocarcinomas demonstrated 23 (11%) cases with the mutant allele peak higher than the wild-type allele peak and 15 (7%) cases with the mutant allele peak equal to the wild-type allele peak. Of 17 cases with the mutant allele peak higher or equal to the wild-type allele peak, 8 (47%) showed KRAS amplification by FISH. KRAS FISH analysis of 36 KRAS-mutated lung adenocarcinomas with the mutant allele peak lower than the wild-type allele peak, 21 KRAS and EGFR wild-type and 16 EGFR-mutated adenocarcinomas showed no KRAS amplification. KRAS MASI was associated with selective amplification of the KRAS mutant allele (P<0.001). Patients with KRAS MASI showed worse overall survival. The cumulative proportion surviving at 17 months for KRAS MASI group was 35% compared with 84.1% for patients with KRAS mutant allele peak lower than wild-type allele peak (P=0.012). The adverse prognostic significance of KRAS MASI was independent of clinical stage and was maintained among stage I patients. The detection of KRAS MASI in lung adenocarcinomas by sequencing electropherograms may identify patients with more aggressive disease.
Similar content being viewed by others
Main
About 30% of lung adenocarcinomas in western population harbor KRAS mutations.1, 2 In addition to activating point mutations, the oncogenic potential of KRAS can be achieved by increase in copy number of the KRAS gene.3, 4 Recent studies have shown that in a subset of cases, KRAS mutations were strongly associated with higher KRAS gene copy number.5, 6, 7 The combination of these genetic events may result in an imbalance between the wild-type allele and the mutant allele.
The scenario with mutant allele being predominant over the wild-type allele was defined as mutant allele-specific imbalance (MASI).8, 9, 10 The incomplete dominance of the mutant allele over the wild-type allele may result from selective amplification of the mutant allele (partial MASI) and is likely to be identified by fluorescence in situ hybridization (FISH). In some cases, the mutant allele may be predominant in the absence of the wild-type allele (complete MASI), a phenomenon most likely to arise through acquired uniparental disomy.11
Recently, the combination of KRAS mutation and copy number gain was correlated with worse clinical outcome in lung adenocarcinoma patients.10, 12
KRAS mutation analysis is usually achieved by the automated chain termination method of direct DNA sequencing. When KRAS mutations are present, review of the sequencing electropherogram allows visualization of both the mutant allele and the wild-type allele. We hypothesized that mutant allele/wild-type allele peak height ratio on sequencing electropherogram is representative of the actual KRAS mutant allele/wild-type allele ratio in the tumor and may offer additional clinical prognostic information in an otherwise relatively homogenous population of KRAS-mutated lung adenocarcinomas. This hypothesis was tested by characterizing KRAS MASI in 207 prospectively accrued lung adenocarcinomas that harbored KRAS mutations. Furthermore, we correlated KRAS MASI with clinicopathological parameters such as KRAS amplification determined by FISH, clinical stage and overall survival.
Materials and methods
Clinicopathological Characteristics of Studied Patients
A total of 244 lung adenocarcinomas prospectively tested for EGFR and KRAS mutations at the University of Pittsburgh Medical Center were included in this study (207 KRAS mutated, 16 EGFR mutated and 21 EGFR/KRAS wild types). The clinicopathological features of the patients with KRAS-mutated adenocarcinomas are summarized in Table 1. There were 124 women and 83 men. All cases were staged according to the seventh edition of the American Joint Committee on Cancer manual.13 Patients with KRAS-positive adenocarcinomas included 201 Caucasians and six African Americans. Only four patients received neoadjuvant chemotherapy before KRAS testing. Information on previous treatment with tyrosine kinase inhibitors was not available. The median follow-up was 17 months. Adequate smoking history was available for 172 patient (65—current smokers, 95—former smokers and 12—never smokers). The high prevalence of smokers in this cohort is consistent with the previously reported high smoking rate among patients treated at the University of Pittsburgh Medical Center (41 never smokers and 296 smokers).14 This study was approved by the University of Pittsburgh Medical Center Institutional Review Board (IRB no. PRO08040162).
KRAS and EGFR Mutation Analysis
KRAS and EGFR mutation analysis was performed as previously described.15 Briefly, guided by hematoxylin and eosin (H&E)-stained slides, tumor targets containing more than 70% tumor cells were manually microdissected from the 4-μm unstained histological sections. DNA was isolated from each target using the DNeasy tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
For the detection of mutations, DNA was amplified (40 cycles) with primers flanking exon 2 of the KRAS gene (forward primer 5′-GGTGAGTTTGTATTAAAAGGTACTGG-3′ and reverse primer 5′-TCCTGCACCAGTAATATGCA-3′), exon 19 of the EGFR gene (forward primer 5′-CCCAGCAATATCAGCCTTAGGTG-3′ and reverse primer 5′-CCACTAGAGCTAGAAAGGGAAAGAC-3′) and exon 21 of the EGFR gene (forward primer 5′-CCTCACAGCAGGGTCTTCTC-3′ and reverse primer 5′-CCTGGTGTCAGGAAAATGCT-3′). PCR products were sequenced in both the sense and antisense directions using the BigDye Terminator v3.1 cycle sequencing kit on an ABI 3130 automated sequencer (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The sequences were analyzed using Mutation Surveyor software (SoftGenetics, LLC, State College, PA, USA). Cases were classified as mutated or wild type for the KRAS and EGFR mutation based on the sequencing results.
KRAS Fluorescence In Situ Hybridization
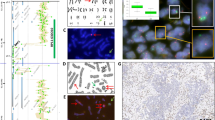
Dual-color KRAS FISH analysis was performed using a Spectrum Green-labeled chromosome enumeration probe 12 (CEP12; Abbott Molecular, Des Plaines, IL) and a Spectrum Orange-labeled, locus-specific KRAS (RP11-295I5, CHORI, Oakland, CA) probe (Figure 1).16 In brief, paraffin sections were deparaffinized, dehydrated in ethanol and air dried. Sections were digested with protease (0.5 mg/ml) at 37°C for 28 min. The slides were denatured at 75°C for 5 min in 70% formamide (Chemicon, Billerica, MA) and dehydrated in ethanol. The probes were denatured for 5 min at 75°C before hybridization. Slides were hybridized overnight at 37°C and washed in 2XSSC/0.3% Igepal (Sigma, St Louis, MO) at 72°C for 2 min. Nuclei were counterstained with DAPI/antifade (Abbott Molecular). Analyses were performed using a fluorescence microscope (Nikon Eclipse E600) and Cytovysion Workstation (Applied Imaging, Santa Clara, CA) equipped with filter sets with single- and dual-band excitors for Spectrum Green, Spectrum Orange and DAPI (UV 360 nm). The histological areas previously selected on the H&E-stained sections were identified on the FISH-treated slides. Only individual and well-delineated cells were scored. Overlapping cells were excluded from the analysis. Amplification was defined as a KRAS/CEP12 ratio of >2.
Representative case of lung adenocarcinoma characterized by fluorescence in situ hybridization (FISH). (a) KRAS/chromosome enumeration probe 12 (CEP12) ratio of >2 indicating KRAS amplification. (b) KRAS FISH-negative control—normal lung tissue showing two CEP12 and two KRAS signals per nucleus. Arrows indicate clusters of amplified KRAS.
Semiquantitative Assessment of KRAS MASI by Comparing Mutant Allele and Wild-Type Allele Peak Heights on Sequencing Electropherograms
Peak heights of mutant and wild-type alleles were compared and grouped into three categories: mutant allele higher than wild-type allele, mutant allele equal to wild-type allele and mutant allele lower than wild-type allele (Figure 2). Any increase in the mutant allele over wild-type allele and cases with equal peaks were defined as MASI.
Scanned segments of sequencing electropherograms of representative cases of lung adenocarcinoma harboring KRAS mutations. The variation in peak height of wild-type allele and mutant allele is illustrated. The top part of each panel shows nucleotide sequence of the dominant PCR product. Point mutation is indicated by an arrow. (a) G to T nucleotide substitution; height of the mutant allele peak (T) is lower than that of the wild-type allele (G). (b) G to A substitution; heights of mutant and wild-type alleles are equal. (c) G to C substitution; mutant allele peak (C) is higher than the wild-type allele peak (G), consistent with partial mutant allele-specific imbalance. (d) G to T substitution; wild-type allele peak is virtually absent, consistent with complete MASI. Of note, this case showed no KRAS amplification or chromosome 12 hyperploidy by FISH, raising the possibility of an acquired uniparental disomy or homozygous mutation.
Statistical Analysis
Overall survival was measured from the date of surgery to the date of death. Only patients who survived for at least 3 months after surgery were included in survival analysis. Living patients were censored at the date of the last available clinical information. Survival probabilities were calculated using the Kaplan–Meier method and compared among different groups and subgroups (log-rank test and Mantel–Cox). Statistical analysis was performed using SPSS 19 (Somers, NY, USA).
Results
Identifying KRAS MASI: Comparing Sequencing Electropherograms and FISH
A review of sequencing electropherograms of 207 KRAS-mutated lung adenocarcinomas identified 15 cases with the mutant allele peak equal to the wild-type allele peak and 23 cases with the mutant allele peak higher than the wild-type allele peak (overall KRAS MASI incidence—38/207, 18%). Sufficient material for KRAS FISH was available in 17 of 38 cases with KRAS MASI. In all, 2 of 7 (29%) KRAS cases with equal peaks and 6 of 10 (60%) KRAS cases with the mutant allele peak higher than the wild-type allele peak showed KRAS amplification. Overall, 8 of 17 (47%) cases with KRAS MASI by sequencing electropherogram showed a KRAS/CEP12 FISH ratio of >2, indicating that amplification of the KRAS mutant allele is a common mechanism of KRAS MASI. None of 36 KRAS cases with the mutant allele peak lower than the wild-type allele peak showed amplification. Therefore, KRAS MASI on sequencing electropherogram is associated with higher incidence of KRAS amplification (0 vs 47%; P<0.001, Fisher exact probability test) (Table 1).
KRAS FISH was also performed on a randomly selected group of KRAS wild-type adenocarcinomas, including 21 KRAS and EGFR wild-type cases and 16 cases harboring EGFR mutations. None of the KRAS and EGFR wild-type or EGFR-mutated cases showed KRAS amplification.
Clinicopathological Correlates of KRAS MASI
The presence of KRAS MASI was not associated with patients' gender, age, smoking history, type of KRAS mutation, tumor morphology or clinical stage (Table 1). Consistent with the previous report, patients with KRAS amplification by FISH were older than patients without KRAS amplification (69 vs 66 years of age);12 however, this difference did not reach statistical significance.
Male patients had worse overall survival; however, this trend did not reach statistical significance (P=0.063). Patients' age, smoking history and type of KRAS mutation did not correlate with overall survival. Patients with KRAS amplification by FISH showed trend to worse overall survival (P=0.182).
The prognostic value of clinical stage is shown in Figure 3a.
Overall survival analysis, Kaplan–Meier method. (a) Overall survival by clinical stage. (b) Overall survival and KRAS mutant allele (MA) to wild-type allele (WA) peak height comparison as seen on sequencing electropherogram. The survival curve for patients with equal mutant and wild-type allele peak heights (MA=WA) resemble that of patients with mutant allele higher than wild-type allele (MA>WA). (c) Overall survival and KRAS mutant allele-specific imbalance (MA⩾WA). (d) To control for clinical stage, the overall survival analysis was performed among patients grouped by clinical stage, stage I through IV. Stage I KRAS MA>WA patients showed worse overall survival. No such relationship was identified for patients with stages II, III or IV.
Overall, this cohort of patients with KRAS-mutated lung adenocarcinoma was quite homogenous, as most of the well-recognized prognostic factors (ie, age and gender) did not correlate with overall survival.
The difference in overall survival was most striking when KRAS-mutated cases with the mutant allele peak higher than the wild-type allele peak were analyzed separately from KRAS-mutated cases with equal peaks (Figure 3b). Among patients with the KRAS mutant allele peak higher than the wild-type allele peak, the cumulative proportion surviving at 17 months was 35%, and for patients with the KRAS mutant allele peak lower than the wild-type allele peak, it was 84.1% (P=0.012).
The cases with equal height of mutant and wild-type allele peaks were grouped together with cases showing mutant allele peak higher than wild-type allele peak for the following reasons: (1) Two of seven cases with equal mutant and wild-type allele peaks, but none of the cases with the KRAS mutant allele peak lower than the wild-type allele peak showed KRAS amplification; (2) clinical outcome of patients with equal mutant and wild-type allele peaks resembled that of patients with KRAS mutant allele higher than wild-type allele peak (Figure 3b); and (3) combining ‘mutant allele higher than wild-type allele peak’ cases with ‘mutant allele equal to wild-type allele peak’ group allowed statistical subgroup analysis of the possible relationship between clinical stage and KRAS MASI.
Patients with adenocarcinomas characterized by KRAS MASI showed worse overall survival. The mortality among KRAS MASI patients was 12/34 (35.3%) vs 29/158 (18.4%) among patients with KRAS mutant allele peak lower than wild-type allele peak (P=0.07). The cumulative proportion surviving at 18 months was 84.1% in KRAS non-MASI cases and 72.4% in KRAS MASI cases (P=0.003) (Figure 3c).
To determine whether adverse prognostic significance of KRAS MASI is independent of clinical stage, a subgroup analysis was performed. The adverse prognostic effect of KRAS MASI was maintained among patients with stage I disease (Figure 3d). On the other hand, the prognostic value of clinical stage was maintained only among patients with KRAS mutant allele peak lower than wild-type allele peak.
Discussion
The significance of KRAS MASI stems from the fact that the allelic imbalance is maintained after transcription, affects the dosage of KRAS mutant allele and the level of its kinase activity.7, 10 Recently, it was shown that reactivation of p53 in a KRASG12D mouse model of non-small-cell lung carcinoma lead to tumor regression only in tumors with KRAS signal elevated through the amplification of the mutant allele and loss of wild-type allele.17, 18
In this larger series of KRAS-mutated lung adenocarcinomas, we confirm and expand the finding that KRAS MASI correlates with worse clinical outcome. In this clinically homogenous cohort of KRAS-mutated lung adenocarcinomas, clinical stage was the only other prognostic factor that maintained its significance. We believe that the adverse prognostic significance of KRAS MASI is independent of the clinical stage for several reasons. First, cases characterized by KRAS MASI were not associated with more advanced clinical stage. Second, in a subgroup analysis, the presence of KRAS MASI maintained its adverse prognostic value even within stage I patients. Most interestingly, clinical stage predicted outcome only among patients with the KRAS mutant allele peak lower than the wild-type allele peak.
The quantitative nature of the direct sequencing and its reliability in assessing allelic imbalance was previously shown both on cell lines and in clinical tumor samples.7, 10, 19 In addition to sequencing electropherogram and FISH, previous studies of KRAS allelic imbalance in lung adenocarcinomas used such methods as qPCR and SNP (Table 2). The number of clinical samples was lower, and the ethnicity of studied patients was distinct from the present study. Nevertheless, our results are in agreement with the previously reported adverse prognostic impact of the combined KRAS mutation and increased KRAS gene copy number.10, 12
In this study, we employed KRAS FISH to analyze two known mechanisms of KRAS MASI—amplification and chromosome 12 hyperploidy. It appears that these two events are rare and insufficient to explain the adverse prognostic impact of KRAS MASI. The role of two additional MASI mechanisms, chromosome 12 uniparental disomy and KRAS homozygous mutation, should be addressed in future studies.
The reported incidence of KRAS amplification in non-small-cell lung carcinomas ranges from 2.5%12 to 7%16 and its variation among different histological subtypes of non-small-cell lung carcinoma is unclear. The current study was not designed to establish the incidence of KRAS amplification in a general cohort of non-small-cell lung carcinomas—our study was histologically limited to lung adenocarcinomas only. Of the KRAS-mutated adenocarcinomas, KRAS FISH was performed on all cases with KRAS MASI with available material and on additional 36 randomly selected KRAS-mutated adenocarcinomas with the mutant allele peak lower than the wild-type allele peak. This KRAS MASI-centric approach resulted in a 15% (8/53) incidence of KRAS amplification in KRAS-mutated adenocarcinomas and in an 8.8% (8/90) incidence of KRAS amplification in combined KRAS/EGFR wild-type, EGFR- or KRAS-mutated adenocarcinomas.
In summary, KRAS MASI as identified on sequencing electropherograms was seen in 11% (mutant allele peak higher than wild-type allele peak) to 18% (equal mutant and wild-type allele peaks) of 207 patients with KRAS-mutated lung adenocarcinoma. Forty-seven percent of lung adenocarcinomas with KRAS MASI by sequencing electropherogram showed KRAS amplification by FISH. KRAS MASI appears to identify a subset of patients with worse overall survival. On the basis of a subgroup analysis, the adverse prognostic significance of KRAS MASI is independent of clinical stage. The results presented in this study require further validation in larger cohorts including patients from different ethnic groups and with more extensive treatment history.
References
Herbst RS, Heymach JV, Lippman SM . Lung cancer. N Engl J Med 2008;359:1367–1380.
Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008;9:962–972.
Pulciani S, Santos E, Long LK, et al. ras gene Amplification and malignant transformation. Mol Cell Biol 1985;5:2836–2841.
Schwab M, Alitalo K, Varmus HE, et al. A cellular oncogene (c-Ki-ras) is amplified, overexpressed, and located within karyotypic abnormalities in mouse adrenocortical tumour cells. Nature 1983;303:497–501.
Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069–1075.
Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893–898.
Modrek B, Ge L, Pandita A, et al. Oncogenic activating mutations are associated with local copy gain. Mol Cancer Res 2009;7:1244–1252.
Gandhi J, Zhang J, Xie Y, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One 2009;4:e4576.
Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol 2005;23:2513–2520.
Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One 2009;4:e7464.
Engel E . A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet 1980;6:137–143.
Sasaki H, Hikosaka Y, Kawano O, et al. Evaluation of Kras gene mutation and copy number gain in non-small cell lung cancer. J Thorac Oncol 2011;6:15–20.
Edge SB, Byrd DR, Compton CC, et al. AJCC Cancer Staging Manual, 7th edn. Springer: Chicago, 2010.
Dacic S, Shuai Y, Yousem S, et al. Clinicopathological predictors of EGFR/KRAS mutational status in primary lung adenocarcinomas. Mod Pathol 2010;23:159–168.
Chiosea S, Shuai Y, Cieply K, et al. EGFR fluorescence in situ hybridization-positive lung adenocarcinoma: incidence of coexisting KRAS and BRAF mutations. Hum Pathol 2010;41:1053–1060.
Wagner PL, Perner S, Rickman DS, et al. In situ evidence of KRAS amplification and association with increased p21 levels in non-small cell lung carcinoma. Am J Clin Pathol 2009;132:500–505.
Feldser DM, Kostova KK, Winslow MM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 2011;468:572–575.
Junttila MR, Karnezis AN, Garcia D, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 2011;468:567–571.
Oakley GJ, Chiosea SI . Higher Dosage of the Epidermal Growth Factor Receptor Mutant Allele in Lung Adenocarcinoma Correlates with Younger Age, Stage IV at Presentation and Poorer Survival. J Thorac Oncol; advance online publication, 6 May 2011 (e-pub ahead of print).
Acknowledgements
We thank members of the Molecular Anatomic Pathology and FISH and Developmental laboratories of the Department of Pathology, University of Pittsburgh, for excellent technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Chiosea, S., Sherer, C., Jelic, T. et al. KRAS mutant allele-specific imbalance in lung adenocarcinoma. Mod Pathol 24, 1571–1577 (2011). https://doi.org/10.1038/modpathol.2011.109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2011.109
Keywords
This article is cited by
-
Chemical evolution of an autonomous DNAzyme with allele-specific gene silencing activity
Nature Communications (2023)
-
A biologically stable DNAzyme that efficiently silences gene expression in cells
Nature Chemistry (2021)
-
Loss of the wild-type KRAS allele promotes pancreatic cancer progression through functional activation of YAP1
Oncogene (2021)
-
Targeting gallbladder cancer: a pathway based perspective
Molecular Biology Reports (2020)
-
KRAS mutational analysis and immunohistochemical studies can help distinguish pancreatic metastases from primary lung adenocarcinomas
Modern Pathology (2014)