Abstract
Breast adenosquamous carcinomas are rare tumours characterized by well-developed gland formation intimately admixed with solid nests of squamous cells immersed in a highly cellular spindle cell stroma. A low-grade variant has been described that is associated with a better prognosis. Here we studied five cases of adenosquamous carcinomas to determine their genetic profiles and to investigate whether the spindle cell component of these cancers could at least in part stem from the glandular/epithelial components. Five adenosquamous carcinomas of the breast were subjected to (1) immunohistochemical analysis, (2) microdissection and genetic analysis with a high-resolution microarray comparative genomic hybridization platform, and (3) chromogenic in situ hybridization. All cases displayed a triple-negative immunophenotype, consistently expressed ‘basal’ keratins and showed variable levels of epidermal growth factor receptor expression. Microarray comparative genomic hybridization analysis of two of the cases revealed multiple low-level gains and losses affecting several chromosomal arms. Case 1 displayed gains of the whole of chromosome 7, and case 2 harboured a focal, high-level amplification of 7p12, encompassing the epidermal growth factor receptor gene, which was associated with strong and intense membranous epidermal growth factor receptor expression. Chromogenic in situ hybridization revealed that the genetic features found in the epithelial cells were also present in a minority of the spindle cells of the stromal component, in particular in those near the epithelial clusters, indicating that some of the spindle cells are clonal and derived from the epithelial component of the tumour. In conclusion, breast adenosquamous carcinomas are triple-negative cancers that express ‘basal’ keratins. These tumours harbour complex genetic profiles. Some of the spindle cells in adenosquamous carcinomas are derived from the epithelial component, suggesting that adenosquamous carcinomas may also be part of the group of metaplastic breast carcinomas with spindle cell metaplastic elements.
Similar content being viewed by others
Main
Adenosquamous carcinomas of the breast are rare tumours, included in the last edition of the World Health Organization (WHO) classification of breast cancers1, 2 as a subtype of metaplastic carcinoma. They are characterized by well-developed gland/tubule formation intimately admixed with solid nests of squamous cells in a spindle cell background. A low-grade variant was described in 1987 by Rosen and Ernsberger,3 who highlighted that, despite the presence of metaplastic elements, these tumours displayed a low-grade pattern, bearing some resemblance to tubular carcinomas. In agreement with their low-grade morphological features, the majority of low-grade adenosquamous carcinomas have been described to have an excellent prognosis, with a low incidence of lymph node metastasis.1, 4 A proportion of cases, however, can behave in a locally aggressive manner.
At gross examination, low-grade adenosquamous carcinomas tend to display a stellate or infiltrative configuration, with poorly defined borders. Microscopically, the carcinomatous component is characterized by small glandular structures, with rounded rather than angulated contours, and solid cords of epithelial cells, which may contain squamous cells, squamous pearls or squamous cyst formation. The invasive neoplastic component typically shows long, slender extensions at the periphery and infiltrate in between normal breast structures, features which have been associated with inadequate local excision and high incidence of recurrence.1 Clusters of lymphocytes are often observed at the periphery. Furthermore, the association between these tumours and adenomyoepithelioma and sclerosing proliferative lesions has been reported.4, 5
The epithelial structures of adenosquamous carcinomas are often immersed in a highly cellular spindle cell stroma. Stromal cells appear to merge with the epithelial cells and, in fact, in some cases, spindle cell metaplasia is recognized and some have suggested that the stroma should be considered as part of the neoplastic component.1, 3, 4, 5 As emphasized in the first description of the low-grade variant, however, in some instances, it may be ‘difficult to distinguish between spindle cell metaplasia and ordinary stroma’.3 Furthermore, so far, the available evidence in support of the concept that some of the stromal spindle cells are indeed metaplastic, that is, derived from the epithelial cells, remains restricted to phenotypic analyses (ie, histopathological and immunohistochemical). Ultra-structural features of the adenocarcinomatous and squamous cells have been described in detail.4, 6 However, probably due to characteristics of the specimen subjected to electron microscopy, the stroma was reported to consist ‘primarily of numerous collagen fibrils’. Few immunohistochemical studies have described negativity for hormone receptors5, 6, 7 and a high prevalence of HER2 membrane staining (46%) in tumours cells;7 no genome-wide molecular genetic analysis of low-grade adenosquamous carcinomas has been performed to our knowledge.
Here we report on five cases of adenosquamous carcinoma of the breast, which were subjected to high-resolution microarray comparative genomic hybridization, followed by chromogenic in situ hybridization and immunohistochemistry. The aims of this study were twofold: (1) to characterize the genetic profiles of adenosquamous carcinomas and (2) to determine if cells of the spindle cell component harbour the genetic aberrations found in the glandular/epithelial components.
Materials and methods
Cases
Five cases diagnosed as adenosquamous carcinoma of the breast were retrieved from the files of the Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis, IN, USA. Samples were anonymized before analysis. Ethical approval was available.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were cut at 3 μm and mounted on silane-coated slides. Immunohistochemistry was performed as previously described8 using antibodies raised against estrogen receptor (ER), progesterone receptor (PR), HER2 and ‘basal-like’ markers (epidermal growth factor receptor (EGFR), cytokeratin (CK) 5/6, CK14, CK17). The antibody sources, dilutions, pre-treatment protocols and detection methods are listed in Table 1. Positive and negative controls (omission of the primary antibody and IgG-matched serum) were included for each immunohistochemical run. All markers were scored separately in the epithelial/glandular and spindle cell components of the tumours. For ER and PR, only nuclear reactivity was considered specific and the percentage of positive cells was estimated for each marker. For CKs, any cytoplasmic staining in morphologically unequivocal neoplastic cells was recorded as positive. HER2 was evaluated according to current ASCO/CAP guidelines.9 EGFR was scored as previously described.10
Microdissection and DNA Extraction
All tumours were microdissected to ensure >90% of purity of neoplastic cells. Microdissection was performed with a sterile needle under a stereomicroscope (Olympus SZ61, Tokyo, Japan) from 10 consecutive 8-μm-thick sections stained with nuclear fast red as previously described.11 DNA was extracted using the DNeasy Kit (Qiagen, Crawley, UK) according to the manufacturer's recommendations. DNA concentration was measured with the PicoGreen® assay as per the manufacturer's instructions (Invitrogen, Paisley, UK).11
Microarray Comparative Genomic Hybridization
The microarray comparative genomic hybridization platform used for this study was constructed at the Breakthrough Breast Cancer Research Centre and comprises ∼32 000 bacterial artificial chromosome (BAC) clones tiled across the genome. This type of BAC array platform has been shown to be as robust as and to have comparable resolution with high-density oligonucleotide arrays.12, 13, 14 Labelling, hybridization, washes, image acquisition and data normalization were carried out as previously described.11 The final data set comprised 31 367 clones with unambiguous mapping information according to the hg19 of the human genome (http://www.ensembl.org). Copy number changes were categorized as gains, losses or amplifications according to previously validated thresholds for each clone.15 Threshold values were chosen to correspond to three standard deviations of the normal ratios obtained from the filtered clones mapping to chromosomes 1–22, assessed in multiple hybridizations between DNA extracted from a pool of male and female blood donors as previously described16, 17 (Log2 ratio of ±0.12). Low-level gain was defined as a smoothed Log2 ratio of between 0.12 and 0.45, corresponding to approximately 3–5 copies of the locus, whereas gene amplification was defined as having a Log2 ratio >0.45, corresponding to more than five copies.
Chromogenic In Situ Hybridization
Chromogenic in situ hybridization was used to validate the results of microarray comparative genomic hybridization analysis and to investigate whether the detected genetic aberrations were present in the distinct components of the tumours. Ready-to-use digoxigenin-labelled SpotLight amplification probe for EGFR (Zymed, CA, USA) and biotin-labelled centromeric probe for chromosome 7 (CEP7; Zymed) were used. Formalin-fixed, paraffin-embedded sections were cut at 2 μm and mounted on silane-coated slides. Tissue pre-treatment, hybridizations and washes were performed as previously described.18 Signals were counted in the nuclei of 60 morphologically unequivocal tumour cells (epithelial and stromal components) and normal cells for EGFR probe and CEP7. Amplification was defined as the presence of large gene clusters or >5 gene copies in >50% of the neoplastic cells.
Simultaneous Immunohistochemistry and Chromogenic In Situ Hybridization
Formalin-fixed, paraffin-embedded sections were cut at 2 μm and mounted on silane-coated slides. Immunohistochemistry was performed with an antibody against smooth muscle actin (SMA). Details of the antibody source and dilution, and detection method are listed in Table 1. Antibody binding was detected using an alkaline phosphatase-conjugated avidin–biotin complex (ABC) kit (Vector Labs, CA, USA) and Vector Red chromogen (Vector Labs). Slides were incubated for 30 min in secondary antibody and ABC, and for 20 min in chromogen. Immediately after the immunohistochemical reaction, sections were subjected to chromogenic in situ hybridization analysis following the same protocol as described above. Signals were scored as described above.
Results
Cases
All patients were female and predominantly postmenopausal, the age at diagnosis ranging from 54 to 76 years (mean=70; median=72). All but one tumour were staged as pT2, with maximum diameter ranging from 1.5 to 3.7 cm (mean=2.7 cm; median=2.6 cm). Axillary nodal status was available only for one case (case 2), and no metastatic deposits were observed.
Histopathological Analysis
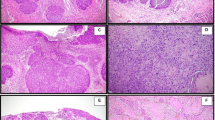
Histopathological review was performed by three of the authors (FG, SB, JR-F). All cases were composed of varying proportions of adenocarcinomatous, squamous and stromal components and four cases (cases 1, 3, 4 and 5) entirely fulfilled the criteria proposed by Rosen and Ernsberger3 for low-grade adenosquamous carcinomas (Figure 1 and Supplementary Figures 1, 2, 3 and 4). These four tumours were characterized by infiltrative borders (Figure 1a) and neoplastic glands tended to infiltrate between ducts and into lobules (Figure 1b); the epithelial component was in the form of small tubules, cell clusters and cords (Figure 1c) with focal squamous differentiation (Figure 1d), haphazardly arranged in an infiltrative spindle cell background. The glandular/epithelial cells tended to subtly merge with the spindle cells (Figure 1e). Lymphocytic aggregates were present in all tumours and conspicuous at the periphery of three cases (Figure 1e). Case 5 was associated with a sclerosing papillary lesion (Figure 1a), which may have possibly been the substrate from which the invasive tumour originated.5 The remaining case (case 2) had areas that displayed the hallmark features of a low-grade adenosquamous carcinoma (Figure 2a and b and Supplementary Figure 5); however, a gradual transition to high-grade features with an overtly malignant spindle cell component was observed (Figure 2a and c and Supplementary Figure 5). This case is consistent with previous description of progression from low-grade adenosquamous carcinomas to a high-grade metaplastic carcinoma.5, 19
Histological features of cases 1, 3, 4 and 5. Tumour borders were infiltrative (a, case 5) and neoplastic cells infiltrated the normal breast structures (b, case 1). Small tubules and cell clusters (c, case 5) with focal squamous differentiation (d, case 4) were arranged in haphazard manner in a spindle cell stroma. The spindle cells often appeared to merge with the epithelial cells (e, case 1). Lymphocytic infiltrate was conspicuous in three cases (f, case 3). Additional representative micrographs of cases 1, 3, 4 and 5 are available in Supplementary Figures 1, 2, 3 and 4, respectively.
Histological features of case 2. A transition from areas typical of low-grade adenosquamous carcinomas (a, at left side, and b) to high-grade areas, which also comprised overtly malignant spindle cells (a, at right side, and c), was observed. Additional representative micrographs of case 2 are available in Supplementary Figure 5.
Immunohistochemistry
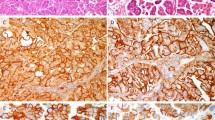
Results of the immunohistochemical analysis are summarized in Table 2 and shown in Figure 3. Immunohistochemical profiles were very similar across the different cases. All tumours were of triple-negative phenotype (ie, ER-, PR- and HER2-negative) (Figure 3a, b and c) with consistent expression of basal markers. Positive reactivity for CK5/6, CK14 (Figure 3d) and CK17 (Figure 3e) was present in all cases, with a diffuse strong staining in the epithelial cells, whereas only scattered stromal cells were positive (Supplementary Figures 1f, 2f, 3f, 4f and 5k and l). EGFR was overexpressed in cases 2, 3 and 5, with predominant staining in the epithelial cells. In case 2, the high-grade component displayed a strong staining for EGFR, including in the stromal spindle cells (Figure 3f). The immunohistochemical profile of all cases was consistent with that of basal-like breast cancers, according to the microarray gene expression profiling-validated immunohistochemical definition proposed by Nielsen et al20 (ie, lack of ER and HER2, and expression of CK5/6 and/or EGFR), which has a specificity of 100% for basal-like breast cancers.
Immunohistochemical features. All tumours were negative for estrogen (a) and progesterone (b) receptors, and HER2 (c). Consistent positivity for basal cytokeratins (CKs), including CK17 (d) and CK14 (e), was observed. EGFR overexpression was present in case 2, with strong staining in spindle cells of the high-grade component (f).
Microarray Comparative Genomic Hybridization
Optimal quality DNA for microarray comparative genomic hybridization experiments was obtained only from cases 1 and 2. In agreement with its low-grade morphological features,21 case 1 displayed a rather simple molecular karyotype (Figure 4a and c), comprising gains of 6p, 7pq and 8q, losses of 1p, 6p, 6q, 8p and 9p, and no amplifications. Case 2 displayed a greater degree of genetic complexity (Figure 4b and b). Its genomic profile was characterized by a complex ‘firestorm’ pattern,22 with two amplification peaks on chromosome 7. Gains of 1q, 5q, 7p, 8q, 12p, 14q, 16p, 16q and 18pq, losses of 1p, 3q, 8p, 9p, 12q, 17p, 17q, 22q and Xpq, and high-level amplification of 7p11.2, encompassing the EGFR gene, and 7q11.21 were observed.
Genome plots and chromosome 7 plots of case 1 (a and c) and case 2 (b and d). Case 1 displayed a rather simple pattern of genomic aberrations, whereas case 2 showed a greater complexity, with a complex firestorm pattern,22 with two regions of high-level amplification in chromosome 7, one of which encompassed the EGFR gene locus. In the genome plots (a and b), CBS (circular binary segmentation) log2 ratios are plotted on the y axis against each clone according to genomic location on the x axis. BACs categorized as displaying genomic gains or amplification are plotted in green and those categorized as genomic losses in red. CBS log2 ratios are plotted in purple. In the chromosome 7 plots (c and d), CBS log2 ratios are plotted on the x axis against each clone according to genomic location on the y axis. BACs categorized as displaying genomic gains or amplification are plotted in green and those categorized as genomic losses in red.
Chromogenic In Situ Hybridization
To validate microarray comparative genomic hybridization findings and study the presence of the detected copy number changes in the distinct components of the tumours, we performed chromogenic in situ hybridization for selected regions harbouring gains or amplification. Microarray comparative genomic hybridization analysis revealed that in case 1 the whole of chromosome 7 harboured gains (Figure 4c). Chromogenic in situ hybridization for the EGFR gene, which maps to 7p11.2, and CEP7 confirmed this finding, showing averages EGFR and CEP7 copy numbers consistent with gain of a copy of chromosome 7 in the epithelial cell clusters and also in the surrounding stromal tumour cells (Table 3). Microarray comparative genomic hybridization analysis of case 2 revealed gains of 7p-q11.21, encompassing the centromere and a high-level amplification of EGFR (Figure 4d). In this case, chromogenic in situ hybridization showed that not only the epithelial cells, but also a minority of the spindle cells in the stromal component, in particular those near the epithelial clusters, harboured EGFR amplification (Figure 5a and b, Table 3). It should be noted that spindle cells harbouring EGFR amplification were found both in the low- and high-grade areas of this tumour (Figure 5a and Supplementary Figures 5q to v). In addition, CEP7 chromogenic in situ hybridization showed gains of the centromeric region in the epithelial and surrounding stromal cells (Table 3).
Chromogenic in situ hybridization (a and b) and simultaneous immunohistochemistry and chromogenic in situ hybridization (c) of case 2. Chromogenic in situ hybridization was performed with a probe for the EGFR gene, and low-grade (a) and high-grade (b) areas are depicted. In c, simultaneous immunohistochemistry and chromogenic in situ hybridization were performed with antibodies against smooth muscle actin and a probe for EGFR, respectively. Note that clusters of epithelial cells and some of the spindle cells (arrows and insets in a and b) of the stromal component display large EGFR gene clusters. Note that other stromal cells display only two copies of EGFR. In addition, spindle-shaped cells (arrow, c) display large EGFR gene clusters (brown) and express actin (red).
Simultaneous Immunohistochemistry and Chromogenic In Situ Hybridization
To further show that some of the stromal spindle cells in case 2 harboured the same genetic aberrations identified in the glandular/epithelial components, we performed simultaneous immunohistochemistry with an antibody against SMA and chromogenic in situ hybridization with EGFR probe. As expected, the great majority of epithelial/glandular cells displayed large gene clusters and did not express SMA (Figure 5c, right side). However, a minority of the spindle cells displayed large gene clusters and expressed SMA (Figure 5c, left side), indicating that neoplastic epithelial cells underwent some degree of epithelial-to-mesenchymal transition (EMT) and acquired a spindle cell phenotype.
Discussion
Adenosquamous carcinoma of the breast is an uncommon histological type of breast cancer, characterized by the presence of well-defined adenocarcinomatous and squamous components. It is accepted that spindle cell metaplasia may occur and a neoplastic spindle cell component coexist with the epithelial components. A low-grade variant has been recognized, which displays a distinct morphology and a better clinical outcome.1, 7 These tumours are still poorly characterized at the molecular level. Here we have described an immunohistochemical and genetic analyses of five cases of adenosquamous carcinoma, four of which were bona fide examples of the low-grade variant.
All tumours studied here were of triple-negative phenotype. Negativity for hormone receptors has been previously reported;5, 7 however, in contrast with the results of Drudis et al,7 who reported that 46% of these tumours displayed HER2 overexpression, the five adenosquamous carcinomas studied here consistently lacked HER2 expression and the two cases subjected to microarray comparative genomic hybridization analysis lacked HER2 gene amplification. These discrepant results are probably due to different scoring methods, given that at that moment7 HER2 status evaluation was not performed according to the current clinical guidelines.9 Expression of basal markers was found in all tumours. The immunoprofile of adenosquamous carcinomas falls into the basal-like molecular subtype according to an immunohistochemical surrogate,20 which has a specificity of 100% for the identification of basal-like breast cancers defined by microarray gene expression profiling. This is in agreement with the observation that focal squamous metaplasia in invasive ductal carcinomas is strongly associated and predictive of the basal-like phenotype23, 24 and with the fact that >90% of metaplastic breast cancers are of basal-like phenotype by immunohistochemical25, 26 and microarray-based analysis.27, 28 Moreover, our results highlight the heterogeneity of basal-like2, 29, 30 and triple-negative31, 32 cancers: although as a group triple-negative and basal-like cancers are associated with a more aggressive clinical behaviour,33, 34, 35 they also encompass rare entities associated with a good prognosis.27, 29, 30, 31, 36, 37
By performing microarray comparative genomic hybridization followed by chromogenic in situ hybridization in two of the cases, we provide direct molecular evidence that a proportion of the spindle cells of the stromal component of adenosquamous carcinomas harbour the same genetic aberrations present in the glandular/epithelial cells (Figure 5 and Table 3) and are indeed derived from the epithelial cells. To corroborate this finding, we have shown that, in case 2, the spindle-shaped cells harbouring EGFR amplification expressed SMA (Figure 5c). Consistent with this observation EGFR protein overexpression was observed in the overtly malignant spindle cells of the high-grade areas of this case (Figure 3f). Furthermore, a minority of the spindle cells surrounding the epithelial clusters expressed CKs (Figure 3e). Taken together, our results provide direct evidence to support the hypothesis that at least part of the so-called stromal component of adenosquamous carcinoma is in fact derived from the epithelial/glandular component and that some form of EMT may take place in adenosquamous carcinomas, in a way akin to the EMT described in other forms of metaplastic breast cancer and mixed malignant Mullerian tumours.27, 38, 39
In conclusion, we have shown that adenosquamous carcinomas of the breast, including the low-grade variant, harbour complex genomic profiles, are of triple-negative phenotype, express basal makers and may harbour EGFR gene amplification. Furthermore, we have provided direct molecular evidence that a minority of the spindle cells of the stromal component are derived from the epithelial cells, suggesting that some form of EMT may take place in this subtype of metaplastic carcinoma. It should be noted that it is not uncommon for adenosquamous carcinomas to have genuine sarcomatoid stroma, as previously described5, 19 and also observed in the cases described here (Figure 2c). Furthermore, the presence of CK-positive spindle cells in cases of low-grade adenosquamous carcinomas is not uncommon.5 Taken together, these lines of evidence suggest that low-grade adenosquamous carcinomas may be best classified as tumours displaying squamous and spindle cell metaplasia rather than ‘pure epithelial squamous cell carcinomas’, as described in the current WHO classification.1, 2
References
Tavassoli FA, Devilee P, (eds). Tumours of the Breast. International Agency for Research of Cancer (IARC): Lyon, France, 2003.
Weigelt B, Reis-Filho JS . Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol 2009;6:718–730.
Rosen PP, Ernsberger D . Low-grade adenosquamous carcinoma. A variant of metaplastic mammary carcinoma. Am J Surg Pathol 1987;11:351–358.
Van Hoeven KH, Drudis T, Cranor ML, et al. Low-grade adenosquamous carcinoma of the breast. A clinocopathologic study of 32 cases with ultrastructural analysis. Am J Surg Pathol 1993;17:248–258.
Denley H, Pinder SE, Tan PH, et al. Metaplastic carcinoma of the breast arising within complex sclerosing lesion: a report of five cases. Histopathology 2000;36:203–209.
Woodard BH, Brinkhous AD, McCarty Sr KS, et al. Adenosquamous differentiation in mammary carcinoma: an ultrastructural and steroid receptor study. Arch Pathol Lab Med 1980;104:130–133.
Drudis T, Arroyo C, Van Hoeven K, et al. The pathology of low-grade adenosquamous carcinoma of the breast. An immunohistochemical study. Pathol Annu 1994;29 (Part 2):181–197.
Tan DS, Marchio C, Jones RL, et al. Triple negative breast cancer: molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res Treat 2008;111:27–44.
Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007;25:118–145.
Reis-Filho JS, Pinheiro C, Lambros MB, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol 2006;209:445–453.
Marchio C, Iravani M, Natrajan R, et al. Genomic and immunophenotypical characterization of pure micropapillary carcinomas of the breast. J Pathol 2008;215:398–410.
Coe BP, Ylstra B, Carvalho B, et al. Resolving the resolution of array CGH. Genomics 2007;89:647–653.
Gunnarsson R, Staaf J, Jansson M, et al. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia—a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer 2008;47:697–711.
Tan DS, Lambros MB, Natrajan R, et al. Getting it right: designing microarray (and not ‘microawry’) comparative genomic hybridization studies for cancer research. Lab Invest 2007;87:737–754.
Reis-Filho JS, Drury S, Lambros MB, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet 2008;40:809–810; author reply 10–12.
Arriola E, Marchio C, Tan DS, et al. Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest 2008;88:491–503.
Simpson PT, Reis-Filho JS, Lambros MB, et al. Molecular profiling pleomorphic lobular carcinomas of the breast: evidence for a common molecular genetic pathway with classic lobular carcinomas. J Pathol 2008;215:231–244.
Lambros MB, Simpson PT, Jones C, et al. Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest 2006;86:398–408.
Rosen P, (ed). Rosen's Breast Pathology, 2nd edn. Lippincott Williams & Wilkins: Philadelphia, PA, 2001.
Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367–5374.
Geyer FC, Lopez-Garcia MA, Lambros MB, et al. Genetic characterisation of breast cancer and implications for clinical management. J Cell Mol Med 2009.
Hicks J, Krasnitz A, Lakshmi B, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 2006;16:1465–1479.
Fulford LG, Easton DF, Reis-Filho JS, et al. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology 2006;49:22–34.
Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol 2006;19:264–271.
Reis-Filho JS, Milanezi F, Steele D, et al. Metaplastic breast carcinomas are basal-like tumours. Histopathology 2006;49:10–21.
Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol 2010;220:562–573.
Weigelt B, Kreike B, Reis-Filho JS . Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res Treat 2009;117:273–280.
Weigelt B, Horlings HM, Kreike B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol 2008;216:141–150.
Rakha EA, Reis-Filho JS, Ellis IO . Basal-like breast cancer: a critical review. J Clin Oncol 2008;26:2568–2581.
Weigelt B, Baehner FL, Reis-Filho JS . The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol 2010;220:263–280.
Reis-Filho JS, Tutt AN . Triple negative tumours: a critical review. Histopathology 2008;52:108–118.
Turner N, Lambros MB, Horlings HM, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 2010.
Cheang MC, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 2008;14:1368–1376.
Rakha EA, Elsheikh SE, Aleskandarany MA, et al. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 2009;15:2302–2310.
Weigelt B, Mackay A, A’Hern R, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol 2010;11:339–349.
Fadare O, Tavassoli FA . The phenotypic spectrum of basal-like breast cancers: a critical appraisal. Adv Anat Pathol 2007;14:358–373.
Gusterson B . Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer 2009;9:128–134.
Sarrio D, Rodriguez-Pinilla SM, Hardisson D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 2008;68:989–997.
Lien HC, Hsiao YH, Lin YS, et al. Molecular signatures of metaplastic carcinoma of the breast by large-scale transcriptional profiling: identification of genes potentially related to epithelial-mesenchymal transition. Oncogene 2007;26:7859–7871.
Acknowledgements
This study was funded by Breakthrough Breast Cancer.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Geyer, F., Lambros, M., Natrajan, R. et al. Genomic and immunohistochemical analysis of adenosquamous carcinoma of the breast. Mod Pathol 23, 951–960 (2010). https://doi.org/10.1038/modpathol.2010.82
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2010.82
Keywords
This article is cited by
-
Metaplastic carcinoma of the breast with squamous differentiation: prognostic factors and multidisciplinary treatment
World Journal of Surgical Oncology (2022)
-
Breast carcinomas of low malignant potential
Virchows Archiv (2022)
-
Adenosquamous carcinoma of the breast: a population-based study
Breast Cancer (2021)
-
Human Metaplastic Breast Carcinoma and Decorin
Cancer Microenvironment (2017)
-
Cytogenetic analysis of metaplastic squamous cell carcinoma of the breast inter- and intratumoral heterogeneity
Breast Cancer (2017)